Abstract
High-grade gliomas (HGG) are a devastating group of cancers, and represent the leading cause of brain tumour-related death in both children and adults. Therapies aimed at mechanisms intrinsic to glioma cells have translated to only limited success; effective therapeutic strategies will need also to target elements of the tumour microenvironment that promote glioma progression. Neuronal activity promotes the growth of a range of molecularly and clinically distinct HGG types, including adult and paediatric glioblastoma (GBM), anaplastic oligodendroglioma, and diffuse intrinsic pontine glioma (DIPG)1. An important mechanism that mediates this neural regulation of brain cancer is activity-dependent cleavage and secretion of the synaptic adhesion molecule neuroligin-3 (NLGN3), which promotes glioma proliferation through the PI3K–mTOR pathway1. However, the necessity of NLGN3 for glioma growth, the proteolytic mechanism of NLGN3 secretion, and the further molecular consequences of NLGN3 secretion in glioma cells remain unknown. Here we show that HGG growth depends on microenvironmental NLGN3, identify signalling cascades downstream of NLGN3 binding in glioma, and determine a therapeutically targetable mechanism of secretion. Patient-derived orthotopic xenografts of paediatric GBM, DIPG and adult GBM fail to grow in Nlgn3 knockout mice. NLGN3 stimulates several oncogenic pathways, such as early focal adhesion kinase activation upstream of PI3K–mTOR, and induces transcriptional changes that include upregulation of several synapse-related genes in glioma cells. NLGN3 is cleaved from both neurons and oligodendrocyte precursor cells via the ADAM10 sheddase. ADAM10 inhibitors prevent the release of NLGN3 into the tumour microenvironment and robustly block HGG xenograft growth. This work defines a promising strategy for targeting NLGN3 secretion, which could prove transformative for HGG therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Venkatesh, H. S. et al. Neuronal activity promotes glioma growth through neuroligin-3 secretion. Cell 161, 803–816 (2015)
Varoqueaux, F. et al. Neuroligins determine synapse maturation and function. Neuron 51, 741–754 (2006)
Radyushkin, K. et al. Neuroligin-3-deficient mice: model of a monogenic heritable form of autism with an olfactory deficit. Genes Brain Behav. 8, 416–425 (2009)
Tabuchi, K. et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science 318, 71–76 (2007)
Etherton, M. et al. Autism-linked neuroligin-3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proc. Natl Acad. Sci. USA 108, 13764–13769 (2011)
Blundell, J. et al. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J. Neurosci. 30, 2115–2129 (2010)
Ni, J. et al. Combination inhibition of PI3K and mTORC1 yields durable remissions in mice bearing orthotopic patient-derived xenografts of HER2-positive breast cancer brain metastases. Nat. Med. 22, 723–726 (2016)
Ozawa, T. et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 26, 288–300 (2014)
Martinho, O. et al. Expression, mutation and copy number analysis of platelet-derived growth factor receptor A (PDGFRA) and its ligand PDGFA in gliomas. Br. J. Cancer 101, 973–982 (2009)
Sakakini, N. et al. A positive feed-forward loop associating EGR1 and PDGFA promotes proliferation and self-renewal in glioblastoma stem cells. J. Biol. Chem. 291, 10684–10699 (2016)
Jung, E. et al. Tweety-homolog 1 drives brain colonization of gliomas. J. Neurosci. 37, 6837–6850 (2017)
Nagaraja, S. et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell 31, 635–652.e6 (2017)
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014)
Bergles, D. E., Roberts, J. D., Somogyi, P. & Jahr, C. E. Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature 405, 187–191 (2000)
Lin, S. C. & Bergles, D. E. Synaptic signaling between GABAergic interneurons and oligodendrocyte precursor cells in the hippocampus. Nat. Neurosci. 7, 24–32 (2004)
Ziskin, J. L., Nishiyama, A., Rubio, M., Fukaya, M. & Bergles, D. E. Vesicular release of glutamate from unmyelinated axons in white matter. Nat. Neurosci. 10, 321–330 (2007)
Suzuki, K. et al. Activity-dependent proteolytic cleavage of neuroligin-1. Neuron 76, 410–422 (2012)
Peixoto, R. T. et al. Transsynaptic signaling by activity-dependent cleavage of neuroligin-1. Neuron 76, 396–409 (2012)
Kuhn, P.-H. et al. Systematic substrate identification indicates a central role for the metalloprotease ADAM10 in axon targeting and synapse function. eLife 5, 1174–1189 (2016)
Lundgren, J. L. et al. ADAM10 and BACE1 are localized to synaptic vesicles. J. Neurochem. 135, 606–615 (2015)
Qu, M., Qiu, B. O., Xiong, W., Chen, D. & Wu, A. Expression of a-disintegrin and metalloproteinase 10 correlates with grade of malignancy in human glioma. Oncol. Lett. 9, 2157–2162 (2015)
Bulstrode, H. et al. A-Disintegrin and Metalloprotease (ADAM) 10 and 17 promote self-renewal of brain tumor sphere forming cells. Cancer Lett. 326, 79–87 (2012)
Siney, E. J. et al. Metalloproteinases ADAM10 and ADAM17 mediate migration and differentiation in glioblastoma sphere-forming cells. Mol. Neurobiol. 54, 3893–3905 (2016)
Kohutek, Z. A., diPierro, C. G., Redpath, G. T. & Hussaini, I. M. ADAM-10-mediated N-cadherin cleavage is protein kinase C-alpha dependent and promotes glioblastoma cell migration. J. Neurosci. 29, 4605–4615 (2009)
Infante J, Burris HA, L. N. et al. A multicenter phase Ib study of the safety, pharmacokinetics, biological activity and clinical efficacy of INCB7839, a potent and selective inhibitor of ADAM10 and ADAM17. Breast Cancer Res. Treat. 106 (Suppl.), S269 (2007)
Friedman, S. et al. Clinical benefit of INCB7839, a potent and selective inhibitor of ADAM10 and ADAM17, in combination with trastuzumab in metastatic HER2 positive breast cancer patients. Cancer Res. 69, 5056 (2014)
Postina, R. et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J. Clin. Invest. 113, 1456–1464 (2004)
Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009)
Rush, J. et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 (2005)
Stokes, M. P. et al. Complementary PTM profiling of drug response in human gastric carcinoma by immunoaffinity and IMAC methods with total proteome analysis. Proteomes 3, 160–183 (2015)
Stokes, M. P. et al. PTMScan direct: identification and quantification of peptides from critical signaling proteins by immunoaffinity enrichment coupled with LC–MS/MS. Mol. Cell. Proteomics 11, 187–201 (2012)
Olsen, J. V. et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 (2005)
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994)
Huttlin, E. L. et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010)
Villén, J., Beausoleil, S. A., Gerber, S. A. & Gygi, S. P. Large-scale phosphorylation analysis of mouse liver. Proc. Natl Acad. Sci. USA 104, 1488–1493 (2007)
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013)
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
Huang, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols 4, 44–57 (2009)
Huang, W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009)
Franklin, K. & Paxinos, G. The Mouse Brain in Stereotaxic Coordinates 3rd edn (Academic, 2008)
Grasso, C. S. et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat. Med. 21, 827 (2015)
Verhaak, R. G. W. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010)
Acknowledgements
The authors gratefully acknowledge support from the V Foundation, Liwei Wang Research Fund, National Institutes of Health (NINDS R01NS092597; NCI 1F31CA200273, P50 CA168504, P50 CA165962, R35 CA210057), Department of Defense (W81XWH-151-0131), McKenna Claire Foundation, Alex’s Lemonade Stand Foundation, The Cure Starts Now Foundation and DIPG Collaborative, Lyla Nsouli Foundation, Unravel Pediatric Cancer, California Institute for Regenerative Medicine (RN3-06510), Childhood Brain Tumor Foundation, Matthew Larson Foundation, the Joey Fabus Childhood Cancer Foundation, the Wayland Villars DIPG Foundation, the Connor Johnson, Zoey Ganesh, and Declan Gloster Memorial Funds, N8 Foundation, Virginia and D.K. Ludwig Fund for Cancer Research, Child Health Research Institute at Stanford Anne T. and Robert M. Bass Endowed Faculty Scholarship in Pediatric Cancer and Blood Diseases, and Breast Cancer Research Foundation and the intramural programs of the National Center for Advancing Translational Sciences and the National Cancer Institute.
Author information
Authors and Affiliations
Contributions
H.S.V., L.T.T., P.J.W., S.M.G., J.L., D.Y.D. and P.J.M. conducted experiments. H.S.V. and M.M. designed the experiments. H.S.V., S.N., S.M.G., J.L., C.J.T. and M.M. analysed the data. J.N. and J.J.Z. developed the breast cancer brain metastasis xenograft model. All authors contributed to manuscript editing. H.S.V. and M.M. wrote the manuscript. M.M. supervised all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
M.M. and H.S.V. declare that Stanford University filed a patent application (15/011260) related to this work.
Additional information
Reviewer Information Nature thanks G. Murphy, M. Taylor and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
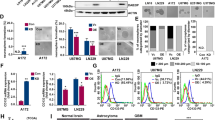
Extended Data Figure 1 Orthotopic xenografts of paediatric glioblastoma fail to grow in the NLGN3-deficient brain.
a, Engraftment is equivalent in Nlgn3 knockout and wild-type mice. In vivo bioluminescence imaging of SU-pcGBM2 xenografts 2 weeks after xenograft in WT;NSG (WT; left) or Nlgn3 KO;NSG mice (KO; right). The heat map superimposed over the mouse heads represents the degree of photon emission by cells expressing firefly luciferase. b, Absolute flux of pHGG cells in identically manipulated WT;NSG (n = 11) and Nlgn3 KO;NSG (n = 14) mice, measured by IVIS imaging 2 weeks after xenograft illustrates no significant difference (P > 0.05) in tumour engraftment (two-sided Mann–Whitney test). Data are mean ± s.e.m. c, d, Data from Fig. 1 shown on the same axis (c) and with each independent cohort colour-coded for comparison of littermates (d). Data illustrate growth of pHGG (SU-pcGBM2) xenografts in identically manipulated WT;NSG (black dots, n = 11) and Nlgn3y/−;NSG (grey dots, n = 14) mice, measured by IVIS imaging (fold change in total photon flux) and shown at 6, 12, 18 and 24 weeks post-xenograft. Data were replicated in five independent cohorts (litters) of mice xenografted with different cell preparations on different days and the data from these five biological replicates are shown combined with each cohort colour-coded (that is, littermates are shown in the same colour). **P < 0.01, ****P < 0.0001, two-sided Mann–Whitney test. Data are mean ± s.e.m.
Extended Data Figure 2 Neuroligin-3 is the only neuroligin family member that promotes glioma proliferation.
a, Schematic representation of active conditioned medium generation (left). Proliferation index (EdU+ and DAPI co-positive nuclei/total DAPI+ nuclei) of pHGG cells (SU-pcGBM2) exposed to plain medium (aCSF), optogenetically stimulated Nlgn3 wild-type cortical slice conditioned medium, or optogenetically stimulated Nlgn3 knockout cortical slice conditioned medium (F = 30.8, P < 0.001). b, c, Proliferation index of patient-derived paediatric cortical glioblastoma (SU-pcGBM2) cells as measured by EdU incorporation 24 h after in vitro exposure to recombinant human NLGN1 at concentrations ranging from 0 to 100 nM (b) or recombinant human NLGN4X or NLGN4Y at 100 nM (c). n = 3 wells per condition. All data are mean ± s.e.m. *P < 0.05, ***P < 0.001, one-way ANOVA with Tukey’s post hoc test for multiple comparisons. n.s. indicates P > 0.05.
Extended Data Figure 3 Gene expression changes induced by neuroligin-3 in glioma.
a, Scatterplot showing SU-pcGBM2 (n = 2) gene expression changes after 16 h of treatment with vehicle (~1% DMSO) or NLGN3 (100 nM). The x axis shows mean fragments per kilobase of transcript per million mapped reads (FPKM) value in vehicle-treated cells, and the y axis shows log2(fold change) of NLGN3 compared to vehicle. Points shown in red represent genes showing statistically significant change (adjusted P < 0.1, Benjaminin–Hochberg for multiple comparison testing). b, Gene Ontology (GO) biological processes enriched in significantly upregulated genes with NLGN3 treatment, as identified by DAVID with P values shown with Benjamini–Hochberg adjustment for multiple comparison testing. c, Genes associated with each GO biological processes terms shown in b.
Extended Data Figure 4 Neuroligin-3 expression data and efficiency of Cre driver mice.
a, Nlgn3 RNA expression (FPKM values) in various cell types (data from brain RNA-seq dataset of Barres and colleagues13). b, c, Recombination rate of inducible Cre driver models 7 days after treatment with tamoxifen (100 mg kg−1 for 5 days) in Rosa26::tdTomatolox-stop-lox reporter mice. b, To assess the neuron-specific CamKIIα–CreER Cre driver, recombination efficiency was quantified as a percentage of NeuN+ neurons that co-express tdTomato+ in the cortex of either CamKIIα–CreER− or CamKIIα–CreER+ mice 7 days after completion of tamoxifen administration. c, To assess the OPC-specific Cre driver PDGFRα–CreER, recombination efficiency was quantified as the number of PDGFRα+ OPCs that co-express tdTomato in the cortex of either PDGFRα–CreER− or PDGFRα–CreER+ mice. n = 3 mice per group.
Extended Data Figure 5 NLGN3 shedding from glioma cells is regulated by NLGN3 exposure and is mediated by ADAM10.
a, NLGN3 western blot illustrating NLGN3 secreted into conditioned medium from optogenetically stimulated Thy1::ChR2; NSG cortical slices (ChR2 stim slice) or SU-DIPG-XIII xenograft-bearing Thy1::ChR2; NSG cortical slices (ChR2 stim slice with xenograft). Performed in biological duplicate. b, NLGN3 western blot illustrating NLGN3 secreted into conditioned medium from wild-type brain slices, wild-type brain slices bearing xenografts of adult GBM SU-GBM035 (WT + xeno), or from Nlgn3 knockout brains bearing SU-GBM035 xenografts (Nlgn3 KO + xeno) in the absence (left three lanes) or presence (right three lanes) of 200 nM ADAM10 inhibitor GI254023X (+ADAM10i). Performed in biological triplicate. c, NLGN3 western blot illustrating glioma cell secretion of NLGN3 in vitro at baseline medium conditions (aCSF), following exposure to recombinant NLGN3 with subsequent washing (NLGN3), at baseline medium conditions in the presence of ADAM10 inhibitor GI254023X (aCSF + ADAM10i) or after NLGN3 exposure in the presence of ADAM10 inhibitor (NLGN3 + ADAM10i). Performed in biological triplicate. d, mRNA expression levels of ADAM10 in primary tumour and cultures of DIPG by RNA-seq with values reported as FPKM12,42 (left; n = 8 primary samples, n = 7 culture samples) and in 493 individual adult glioblastoma samples from TCGA43 (right). Values are reported as robust multi-array averages (RMA; right). Boxes show the median, 25th and 75th percentiles, error bars show the minima and maxima.
Extended Data Figure 6 Functional consequences for glioma of protease inhibition in situ and in vitro.
a, SU-pcGBM2 cells (EdU, red; DAPI, blue) exposed to conditioned medium generated in the presence or absence of ADAM10 inhibitor. Scale bar, 50 μm. b–d, Proliferation indices of SU-pcGBM2 cells exposed to plain medium (aCSF) or active conditioned medium generated in the presence or absence of pan-MMP inhibitor (BAT) (b), ADAM10 inhibitor (c) or ADAM10 inhibitor with or without NLGN3 rescue (d). n = 3 wells per condition. e, Cell viability of SU-pcGBM2 cells exposed to ADAM10 inhibitor (GI254023X, 10 nM–2 μM) at 24, 48 and 72 h (n = 3 wells per condition). f, Proliferation index of SU-pcGBM2 cells exposed to GI254023X (0–2 μM) (n = 3 wells per condition). g, Spheroid invasion index of SU-DIPG-VI cells exposed to ADAM10i (0–5 μM) at 24, 48 and 72 h expressed as the diameter of the sphere of glioma cells relative to the initial diameter at time 0 h. h, Neurosphere formation assay in SU-pcGBM2 cells in the presence of GI254023X (0–2 μM; n = 10 wells per condition. i, Extreme limiting dilution assay (ELDA) data presented in h re-plotted here as a log fraction plot with the slope of the solid line representing the log-active cell fraction and confidence intervals shown as dotted lines. SU-pcGBM2 cells treated with ADAM10 inhibitor GI254023X at 0.5 μM (black), 1 μM (red) or 2 μM (green), with vehicle (DMSO) control (royal blue) or no DMSO (cyan) and analysed for neurosphere formation at 2 weeks. In b–d and f, P values as indicated; one-way ANOVA with Tukey’s post hoc test for multiple comparisons. Data are mean ± s.e.m. In h, χ2 test; data are mean ± confidence intervals. n.s. indicates P > 0.05.
Extended Data Figure 7 Glioma xenograft proliferation after ADAM10 inhibition and pharmacokinetics of XL-784.
a, Representative confocal images (Ki67, green; human nuclear antigen, red; MBP, white) of vehicle-treated or ADAM10i-treated mice bearing frontal cortex SU-pcGBM2 xenografts; images similar to but lower magnification than those shown in Fig. 4e; n = 4 vehicle, n = 4 ADAM10i-treated mice. Scale bar, 100 μm. b, Brain tissue and plasma levels of XL-784 at various time points following a single 50 mg kg−1 intraperitoneal dose in NSG mice as assessed by LC–MS/MS. n = 3 mice at each data point. Data are mean ± s.d.
Extended Data Figure 8 Lack of detectable neurotoxicity after treatment with INCB7839.
Histological assessment of neuronal integrity was performed in mice treated with INCB7839 or vehicle control. The cortex, CA1 region of the hippocampus and dentate gyrus of the hippocampus were examined immunohistologically in the hemisphere contralateral to glioma xenografts in mice treated with INCB7839 or vehicle control. Brain sections were immunostained with NeuN (green) to mark neuronal nuclei and cleaved caspase-3 (red) to mark apoptotic cells and counterstained with DAPI (blue). Representative sections from n = 4 INCB7839-treated mice and n = 4 vehicle control mice were examined. Neuronal nuclei appeared morphologically normal and non-pyknotic. Extremely few cleaved caspase-3+ cells were identified in either group; a total of one cleaved caspase-3+ cell was found in each group across all mice examined (white arrows). Scale bar, 200 μm.
Supplementary information
Supplementary Information
This file contains the uncropped western blots and a Supplementary Discussion. (PDF 4235 kb)
Supplementary Data
This file contains phosphoproteomic source data relating to Figure 2 and Extended Data Figure 3. (XLSX 146 kb)
Rights and permissions
About this article
Cite this article
Venkatesh, H., Tam, L., Woo, P. et al. Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature 549, 533–537 (2017). https://doi.org/10.1038/nature24014
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24014
This article is cited by
-
Stabilization of KPNB1 by deubiquitinase USP7 promotes glioblastoma progression through the YBX1-NLGN3 axis
Journal of Experimental & Clinical Cancer Research (2024)
-
A phase Ib/II randomized, open-label drug repurposing trial of glutamate signaling inhibitors in combination with chemoradiotherapy in patients with newly diagnosed glioblastoma: the GLUGLIO trial protocol
BMC Cancer (2024)
-
Synaptic input and Ca2+ activity in zebrafish oligodendrocyte precursor cells contribute to myelin sheath formation
Nature Neuroscience (2024)
-
Glioblastome nutzen neuronale Eigenschaften: Schlüssel zu neuen Therapien?
Der Nervenarzt (2024)
-
Integrating Machine Learning and Mendelian Randomization Determined a Functional Neurotrophin-Related Gene Signature in Patients with Lower-Grade Glioma
Molecular Biotechnology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.