Abstract
Metabolic production of acetyl coenzyme A (acetyl-CoA) is linked to histone acetylation and gene regulation, but the precise mechanisms of this process are largely unknown. Here we show that the metabolic enzyme acetyl-CoA synthetase 2 (ACSS2) directly regulates histone acetylation in neurons and spatial memory in mammals. In a neuronal cell culture model, ACSS2 increases in the nuclei of differentiating neurons and localizes to upregulated neuronal genes near sites of elevated histone acetylation. A decrease in ACSS2 lowers nuclear acetyl-CoA levels, histone acetylation, and responsive expression of the cohort of neuronal genes. In adult mice, attenuation of hippocampal ACSS2 expression impairs long-term spatial memory, a cognitive process that relies on histone acetylation. A decrease in ACSS2 in the hippocampus also leads to defective upregulation of memory-related neuronal genes that are pre-bound by ACSS2. These results reveal a connection between cellular metabolism, gene regulation, and neural plasticity and establish a link between acetyl-CoA generation ‘on-site’ at chromatin for histone acetylation and the transcription of key neuronal genes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Kandel, E. R., Dudai, Y. & Mayford, M. R. The molecular and systems biology of memory. Cell. 157, 163–186 (2014)
Zovkic, I. B., Guzman-Karlsson, M. C. & Sweatt, J. D. Epigenetic regulation of memory formation and maintenance. Learn. Mem. 20, 61–74 (2013)
Gräff, J. & Tsai, L.-H. Histone acetylation: molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 14, 97–111 (2013)
Wood, M. A. et al. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn. Mem. 12, 111–119 (2005)
Korzus, E., Rosenfeld, M. G. & Mayford, M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 42, 961–972 (2004)
Kaelin, W. G. Jr & McKnight, S. L. Influence of metabolism on epigenetics and disease. Cell. 153, 56–69 (2013)
Katada, S., Imhof, A. & Sassone-Corsi, P. Connecting threads: epigenetics and metabolism. Cell. 148, 24–28 (2012)
Cai, L., Sutter, B. M., Li, B. & Tu, B. P. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell. 42, 426–437 (2011)
Wellen, K. E. et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 324, 1076–1080 (2009)
Gut, P. & Verdin, E. The nexus of chromatin regulation and intermediary metabolism. Nature. 502, 489–498 (2013)
Pietrocola, F., Galluzzi, L., Bravo-San Pedro, J. M., Madeo, F. & Kroemer, G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 21, 805–821 (2015)
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 445, 168–176 (2007)
Qi, Y., Wang, J. K., McMillian, M. & Chikaraishi, D. M. Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J. Neurosci. 17, 1217–1225 (1997)
Comerford, S. A. et al. Acetate dependence of tumors. Cell. 159, 1591–1602 (2014)
Sardi, S. P., Murtie, J., Koirala, S., Patten, B. A. & Corfas, G. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell. 127, 185–197 (2006)
Peleg, S. et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 328, 753–756 (2010)
Aoyama, T. et al. Histone modifiers, YY1 and p300, regulate the expression of cartilage-specific gene, chondromodulin-I, in mesenchymal stem cells. J. Biol. Chem. 285, 29842–29850 (2010)
Vecsey, C. G. et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J. Neurosci. 27, 6128–6140 (2007)
Barrett, R. M. et al. Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology. 36, 1545–1556 (2011)
Gräff, J., Woldemichael, B. T., Berchtold, D., Dewarrat, G. & Mansuy, I. M. Dynamic histone marks in the hippocampus and cortex facilitate memory consolidation. Nat. Commun. 3, 991 (2012)
Ariyannur, P. S. et al. Nuclear-cytoplasmic localization of acetyl coenzyme a synthetase-1 in the rat brain. J. Comp. Neurol. 518, 2952–2977 (2010)
Maren, S. & Holt, W. The hippocampus and contextual memory retrieval in Pavlovian conditioning. Behav. Brain Res. 110, 97–108 (2000)
Stanford, S. C. The Open Field Test: reinventing the wheel. J. Psychopharmacol. 21, 134–135 (2007)
Balderas, I. et al. The consolidation of object and context recognition memory involve different regions of the temporal lobe. Learn. Mem. 15, 618–624 (2008)
Rogers, J. L., Hunsaker, M. R. & Kesner, R. P. Effects of ventral and dorsal CA1 subregional lesions on trace fear conditioning. Neurobiol. Learn. Mem. 86, 72–81 (2006)
Peixoto, L. L. et al. Memory acquisition and retrieval impact different epigenetic processes that regulate gene expression. BMC Genomics. 16, S5 (2015)
Mamiya, N. et al. Brain region-specific gene expression activation required for reconsolidation and extinction of contextual fear memory. J. Neurosci. 29, 402–413 (2009)
Poplawski, S. G. et al. Object-location training elicits an overlapping but temporally distinct transcriptional profile from contextual fear conditioning. Neurobiol. Learn. Mem. 116, 90–95 (2014)
Mashimo, T. et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 159, 1603–1614 (2014)
Gao, X. et al. Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat. Commun. 7, 11960 (2016)
Mariño, G. et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol. Cell. 53, 710–725 (2014)
Takahashi, H., McCaffery, J. M., Irizarry, R. A. & Boeke, J. D. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell. 23, 207–217 (2006)
Walker, D. M., Cates, H. M., Heller, E. A. & Nestler, E. J. Regulation of chromatin states by drugs of abuse. Curr. Opin. Neurobiol. 30, 112–121 (2015)
Shah, P. P. et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 27, 1787–1799 (2013)
Acknowledgements
We thank the NeuronsRUs core of the Mahoney Institute for Neurological Sciences for preparations of primary hippocampal neurons. T.A. is supported by RO1 MH 087463. P.M. and S.L.B. are supported by NIH P01AG031862.
Author information
Authors and Affiliations
Contributions
P.M. and S.L.B. conceived the project. P.M. performed most of the experiments. A.M.D. analysed the CAD RNA-seq datasets. P.M. and G.D. performed and analysed CAD ACSS2i and hippocampal RNA-seq experiments. A.M.D. and G.D. analysed ChIP–seq datasets. P.M. and V.L. performed in vivo ACSS2 knockdown and behavioural characterization. P.M. and S.L.B. wrote the manuscript. All authors reviewed the manuscript and discussed the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 ACSS2 localizes to the nucleus of neurons.
a, Percentage of cells with nuclear staining in ACSS2 immunofluorescence experiments (undiff., undifferentiated CAD cells; diff., differentiated CAD neurons; hippocampal, primary hippocampal neurons day 7; a minimum of 50 cells were examined in three replicate immunofluorescence experiments; t-test undiff. vs diff. P < 0.0001, undiff. vs hippocampal P < 0.0001; error bars, s.e.m.). b, Western blots of cytoplasmic (CE) and nuclear (NE) extracts from undifferentiated CAD cells and differentiated CAD neurons were probed with the indicated antibodies. c, d, Immunofluorescence in primary cortical neurons isolated from C57BL/6 embryos, at day 7 (c) and day 14 (d) of in vitro differentiation culture. ACSS2 locates predominantly to nuclei in differentiated primary cortical neurons. All scale bars, 25 μm. e, Immunofluorescence in primary hippocampal neurons isolated from C57BL/6 embryos at day 14 of in vitro differentiation culture. ACSS2 locates predominantly to nuclei in differentiated primary neurons. f, Immunofluorescence in primary hippocampal neurons at day 7 shows that ACL is chiefly localized to the cytoplasm. g, Neuronal differentiation markers decrease in ACSS2 knockdown cells. CAD cells were infected with lentiviral control (WT) or knockdown vector (shACSS2). Western blots of lysates from stably infected differentiated cells were probed with the indicated antibodies and quantified using ImageJ (n = 3; error bars, s.e.m.).
Extended Data Figure 2 ACSS2 regulates neuronal gene expression.
a, b, Correlation plots of replicate RNA-seq in undifferentiated CAD cells (a) and differentiated CAD neurons (b) for scramble control. c, Transcriptome analysis via RNA-seq, done in two highly correlated biological replicates, identified 894 genes that became upregulated in differentiated CAD neurons (red dots depict genes with >1.6-fold increase). d, Pathway analysis of the 894 upregulated genes (red dots in Fig. 2a) using StringDB. The protein–protein interaction graph depicts a network of binding partners that centres on key players in activity-dependent signalling and synaptic plasticity: Itpr1, Grin1, Nefh, Dync1h1 and Calm1. e, Gene ontology enrichment analysis shows upregulation of neuronal pathways. Gene ontology analysis was used on the 894 genes that become upregulated in differentiated CAD neurons (Extended Data Fig. 2c; identified by RNA-seq, fold-enrichment (FE) > 3.5, FDR < 0.005). f, Genome browser view of Nudt from RNA-seq and ChIP–seq (H4K12ac, H4K5ac, and H3K9ac: mm10 chr5: 140,327,500–140,339,000). g, Relative gene enrichment of H3K9ac, H4K5ac, and H4K12ac at genes that are upregulated during CAD neuron differentiation (>1.6-fold, grey bars) versus all other genes (black bars). h, i, Correlation plots of replicate RNA-seq in undifferentiated CAD cells for ACL knockdown (h), and ACSS2 knockdown (i). j, k, Correlation plots of replicate RNA-seq in differentiated CAD neurons for ACL knockdown (j) and ACSS2 knockdown (k). l, ACL knockdown has a much smaller effect on differentiation-linked upregulation of neuronal gene expression (compare to Fig. 1d). Scatter plot contrasts the fold-change FPKM of induced genes (Extended Data Fig. 2c) between wild-type and ACL knockdown cells. Marginal distributions show histogram and kernel density estimation. Ordinary least squares linear regression is displayed with 95% confidence interval. m, The corresponding quintiles of upregulated genes (red dots in Extended Data Fig. 2c) with the greatest fold-change FPKM increase in wild-type cells. The ACL knockdown showed the same upward trend as the wild-type cells (red bars, compared to black bars in Fig. 1f), contrasting with the severe defect in ACSS2-knockdown cells (green bars; for each quintile, columns represent the mean induction value of genes and error bars represent s.e.m.). n, Box plot of global mRNA transcript levels in undifferentiated and differentiated CAD neurons from RNA-seq in wild-type (scramble control knockdown; grey), ACSS2-knockdown (shACSS2 #25 knockdown; green), and ACL-knockdown (shACL #17 knockdown; red) cells. Genome-wide transcript levels are reduced in differentiated ACL-knockdown cells when compared to differentiated wild-type cells (error bars, s.e.m.), whereas the global reduction in differentiated ACSS2-knockdown cells is less significant when compared to differentiated wild-type cells (error bars, s.d.). o, Genes sensitive to ACSS2 knockdown (top 20%) are also sensitive to ACSS2i treatment, which lowers their expression compared to all genes.
Extended Data Figure 3 ACSS2 is chromatin-bound in differentiated CAD neurons.
a, ChIP–seq in differentiated CAD neurons was performed in replicate with two different antibodies against ACSS2. Correlation plot displays relative enrichment over corresponding MACS peaks (default parameters with input as control, 1,598 peaks). b, Correlation plot displays relative genome-wide ChIP–seq enrichment. c, UCSC Genome Browser views of ChIP–seq tracks show that, upon CAD neuron differentiation, increases in H4K5, H4K12, and H3K9 acetylation over the Nudt1 locus co-occur with ACSS2 enrichment (chr5: 140,326,845–140,339,655). d, UCSC Genome Browser view of indicated ChIP–seq tracks in undifferentiated CAD cells and differentiated CAD neurons over Tab2 locus (chr10: 7,875,000–8,004,000). e, Gene ontology enrichment analysis of the genes most proximate to ACSS2 peaks demonstrates that neuron-specific genes are enriched. f, Frequency of ACSS2 peaks (T antibody) located upstream of their target gene associated with histone acetylation. g, Frequency of ACSS2 peaks (CS antibody) located upstream of their target gene associated with histone acetylation. h, Table shows per cent direct overlap of ACSS2 peaks with H3K9ac, H4K5ac, and H4K12ac broad MACS peaks. i–k, Decile plots depict enrichment of H3K9ac (i), H4K5ac (j), and H4K12ac (k) over ranked deciles of ACSS2 peak enrichment (zeroes removed). l–n, Differentiation-induced co-enrichment of ACSS2 and acetyl broad peaks (MACS). Peak enrichment correlation indicated for H3K9ac (l), H4K5ac (m), and H4K12ac (n). o, Discovered de novo motifs for transcription factor binding sites predicted by HOMER from all ACSS2 ChIP–seq peaks called by MACS in differentiated CAD neurons. p, ChIP–seq enrichment of differentiation-induced genes as a group shows correlation with histone acetylation in differentiated CAD neurons.
Extended Data Figure 4 ACSS2 enrichment co-occurs with histone acetylation at neuronal genes in differentiating CAD neurons.
a, UCSC Genome Browser views of ChIP–seq tracks demonstrate that increases in H4K5, H4K12, and H3K9 acetylation co-occur with ACSS2 enrichment over the Idua (α-l-iduronidase) locus upon CAD neuron differentiation (chr5: 108,649,457–108,687,261). b, At the Slc19A1 (solute carrier family 19 member 1) gene, elevated histone H4K5, H4K12, and H3K9 acetylation levels correspond with increasing ACSS2 enrichment in differentiated CAD neurons (chr10: 76,761,141–77,170,455).
Extended Data Figure 5 Genic ACSS2 enrichment upon CAD neuronal differentiation corresponds to increased histone acetylation.
a–d, Meta-gene enrichment analysis shows ChIP occupancy for ACSS2 (a), H3K9ac (b), H4K5ac (c) and H4K12ac (d) across the top 5% of genes enriched for ACSS2 in differentiated CAD neurons (Top 5% DE; red). The bottom 80% of genes (Bot 80% DE) is shown in blue, and the average signal across all genes (All genes DE) is shown in green. e–h, Meta-gene enrichment analysis shows ChIP occupancy for ACSS2 (e), H3K9ac (f), H4K5ac (g) and H4K12ac (h) at the top 5% of genes that become dynamically bound by ACSS2 upon neuronal differentiation (Top 5% DE; red). The bottom 80% of genes (Bot 80% DE) is shown in blue, and the average signal across all genes (All genes DE) is shown in green. i, Multiple linear regression analysis was implemented to model the interaction between genic ACSS2 enrichment and wild-type gene expression changes, and to visualize the interaction between differentiation-linked gene expression changes and ACSS2 recruitment to chromatin. The contour plot of this fitted regression model displays high levels of ACSS2 enrichment in red and low levels in blue, and is overlaid with the scatter plot of the independent gene expression variables. The visualized model demonstrates that high ACSS2 enrichment corresponds to increased gene expression in differentiated CAD neurons.
Extended Data Figure 6 ACSS2 functions in neuronal histone acetylation.
a, Western blot analysis of whole-cell lysates shows that lentiviral shRNA-mediated knockdown of ACSS2 lowers H3K9 and H3K27 acetylation (compare to Fig. 2g), quantified using ImageJ (n = 3, error bars show s.e.m.). b, Western blot analysis of eluates and supernatants of IgG control and ACSS2 co-immunoprecipitation experiments indicates that ACSS2 binds to acetylated chromatin. c, Western blots of lysates from primary hippocampal neurons (d7) treated for 24 h with the ACSS2i, probed with the indicated antibodies (compare to Fig. 2j), and quantified using ImageJ (n = 3, error bars show s.e.m.).
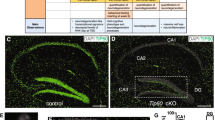
Extended Data Figure 7 ACSS2 chromatin association and H3K9ac in dorsal hippocampus corresponds to H3K27ac and CBP enrichment in neuronal tissue.
a, Genome-wide compartment analysis of in vivo hippocampal ChIP–seq of H3K9ac and mouse forebrain H3K9ac ChIP–seq from ENCODE, showing a similar peak distribution genome-wide: although they originate in different brain regions, the in vivo H3K9ac ChIP data are in strong agreement (Spearman R = 0.67). b, Overlap of RefSeq transcripts targeted by the indicated enzyme or modification (peaks for CBP (GSM1629373) and H3K27ac (GSM1629397) in mouse cortical neurons were called using MACS2 (narrow peaks, FDR 0.1%) with an input sonication efficiency control (GSM1629381); peaks were associated to the nearest TSS among all RefSeq transcripts). c, Gene Ontology enrichment analysis performed on common CBP–ACSS2 targets, indicating that these enzymes co-target genes that modulate synapse biology and synaptic membrane potential.
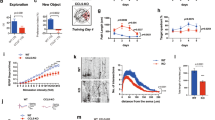
Extended Data Figure 8 Attenuation of ACSS2 expression in the dorsal hippocampus impairs object location memory.
a, ACSS2 RNA in situ hybridization on ACSS2 in sagittal section of hippocampal region CA1 (left, reference atlas adapted from Allen Mouse Brain Atlas12; right, in situ hybridization; HPC, hippocampus proper). b, Weight of eGFP-AAV9 control and shACSS2-AAV9 knockdown mice before intracranial surgery, and following recovery before object location memory (OLM) training (NS, n = 10 per group, error bars show s.d.). c, d, ACSS2 knockdown mice showed no defect in locomotion or thigmotaxis (tendency to remain close to vertical surfaces in an open field, a measure of anxiety), as quantified over 5 min in the open field test; c shows example heat map of tracking data (NS, n = 10 per group, error bars show s.d.). e, Exploration times by eGFP-AAV9 control and shACSS2-AAV9 knockdown mice recorded for the three objects (O1–3) during the first OLM training session (TR) and the 24-h retention test (NL, object in novel location; FL, objects in former location). f, Compared to the control eGFP-AVV9 mice, ACSS2-knockdown mice showed no defect in contextual fear memory. Freezing in chamber on day of contextual fear conditioning was recorded and quantified pre-shock (FC Training; NS, n = 10 per cohort, error bars show s.d.). Fear memory was measured as the freezing response after re-exposure to the context 1 day after contextual fear conditioning (aversive stimulus: 1.5 mA electrical shock; NS, n = 10 per cohort, error bars show s.d.). g, RNA-seq was performed on the dorsal hippocampus of eGFP control and shACSS2-knockdown animals. Global transcript levels were not affected by ACSS2 knockdown (dHPC, dorsal hippocampus; four animals per group, two replicates for each condition; NS, paired t-test, error bars show s.d.). h, Baseline expression of immediate-early genes in untrained animals was unaltered in ACSS2-knockdown mice. RNA-seq was performed on the dorsal hippocampus of eGFP control and shACSS2-knockdown mice (r = 0.82, P < 0.0001; HCC, homecage circadian control).
Extended Data Figure 9 ACSS2 regulates retrieval-induced upregulation of immediate-early genes in vivo.
a, Genome-wide RNA-seq was performed on the dorsal hippocampus of eGFP control and shACSS2-knockdown mice. The analysis was focused on the set of previously identified and validated genes that become upregulated during the sensitive period following memory retrieval. The baseline expression of immediate early genes in untrained animals was not changed in shACSS2-AAV9 mice when compared to eGFP-AAV9 control mice (CC, circadian control). b, During the sensitive period following contextual memory retrieval (RT, 30 min post-exposure to conditioning chamber 24 h after fear conditioning), immediate early genes were upregulated in the dorsal hippocampus of control injected mice. By contrast, the dynamic retrieval-induced expression of these early response genes is absent in ACSS2-knockdown mice (P = 0.001, paired t-test). c, Induction defect of immediate early genes in shACSS2-AAV9 injected animals (RT/CC). d, The baseline expression of genes that were downregulated after contextual memory retrieval is not altered in ACSS2-knockdown mice. e, Downregulation of retrieval-responsive genes occurs in both eGFP control and ACSS2-knockdown mice, except for Cldn5. f, Retrieval-induced downregulation of retrieval-responsive genes in the dorsal hippocampus in eGFP control versus shACSS2-knockdown mice (RT/CC).
Supplementary information
Supplementary Figure 1
This file was replaced on 7 June 2017 to remove a duplicate page. Shown are the original western blots with size marker indications. Outlined in red are the cropped blot data presented in Figures 1c, 1e, 2g, 2h, 2j, 4b and Extended Data Figures 1b and 6b. (PDF 424 kb)
Supplementary Table 1
A list of genes upregulated 1.6-fold or higher upon CAD neuronal differentiation, corresponding to the top 10% of upregulated genes by fold-change. NextSeq mRNA sequencing data were aligned by RNA-STAR 2.3.0.e to the mm10 reference genome, and mapped to genomic features using cufflinks-2.2.1 and mm10 UCSC genomic transcript loci. The rRNA, mRNA, and tRNA of the mouse genome were downloaded from the goldenPath UCSC FTP and were masked from the transcriptome analysis. (TXT 17 kb)
Rights and permissions
About this article
Cite this article
Mews, P., Donahue, G., Drake, A. et al. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546, 381–386 (2017). https://doi.org/10.1038/nature22405
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22405
This article is cited by
-
Histone acetylation in an Alzheimer’s disease cell model promotes homeostatic amyloid-reducing pathways
Acta Neuropathologica Communications (2024)
-
Depletion of SAM leading to loss of heterochromatin drives muscle stem cell ageing
Nature Metabolism (2024)
-
ACSS2 controls PPARγ activity homeostasis to potentiate adipose-tissue plasticity
Cell Death & Differentiation (2024)
-
Microbial short-chain fatty acids regulate drug seeking and transcriptional control in a model of cocaine seeking
Neuropsychopharmacology (2024)
-
Mitochondria in Mesenchymal Stem Cells: Key to Fate Determination and Therapeutic Potential
Stem Cell Reviews and Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.