Abstract
The human X and Y chromosomes evolved from an ordinary pair of autosomes, but millions of years ago genetic decay ravaged the Y chromosome, and only three per cent of its ancestral genes survived. We reconstructed the evolution of the Y chromosome across eight mammals to identify biases in gene content and the selective pressures that preserved the surviving ancestral genes. Our findings indicate that survival was nonrandom, and in two cases, convergent across placental and marsupial mammals. We conclude that the gene content of the Y chromosome became specialized through selection to maintain the ancestral dosage of homologous X–Y gene pairs that function as broadly expressed regulators of transcription, translation and protein stability. We propose that beyond its roles in testis determination and spermatogenesis, the Y chromosome is essential for male viability, and has unappreciated roles in Turner’s syndrome and in phenotypic differences between the sexes in health and disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
BioProject
GenBank/EMBL/DDBJ
Data deposits
The cDNA sequences of Y-linked genes and their X-linked homologs have been deposited in GenBank (http://www.ncbi.nlm.nih.gov) under accession numbers FJ526999–FJ527008, FJ627275, FJ627276, FJ627278, FJ659845, FJ959389, GQ253467–GQ253475, GQ338825, GU304599–GU304603, GU304606, GU304607, JF487792–JF487795, JF827151, JF827152, JN086997, JN585955, JN585956, JQ313990–JQ313992 and BioProject PRJNA221163. The 454 and Illumina testis cDNA sequences have been deposited in GenBank under accession numbers SRX335333, SRX335335, SRX335470, SRX335472, SRX335475–SRX335477, SRX358238 and SRX359414.
References
Lahn, B. T. & Page, D. C. Four evolutionary strata on the human X chromosome. Science 286, 964–967 (1999)
Bellott, D. W. et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466, 612–616 (2010)
Skaletsky, H. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003)
Mueller, J. L. et al. Independent specialization of the human and mouse X chromosomes for the male germline. Nature Genet. 45, 1083–1087 (2013)
Hughes, J. F. et al. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 483, 82–86 (2012)
Hughes, J. F. et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 463, 536–539 (2010)
Hughes, J. F. et al. Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature 437, 100–103 (2005)
Lahn, B. T. & Page, D. C. Functional coherence of the human Y chromosome. Science 278, 675–680 (1997)
Ross, M. T. et al. The DNA sequence of the human X chromosome. Nature 434, 325–337 (2005)
Watson, J. M., Spencer, J. A., Riggs, A. D. & Graves, J. A. The X chromosome of monotremes shares a highly conserved region with the eutherian and marsupial X chromosomes despite the absence of X chromosome inactivation. Proc. Natl Acad. Sci. USA 87, 7125–7129 (1990)
Murtagh, V. J. et al. Evolutionary history of novel genes on the tammar wallaby Y chromosome: implications for sex chromosome evolution. Genome Res. 22, 498–507 (2012)
Hedges, S. B., Dudley, J. & Kumar, S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22, 2971–2972 (2006)
Fisher, R. A. The evolution of dominance. Biol. Rev. Camb. Philos. Soc. 6, 345–368 (1931)
Rozen, S. et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423, 873–876 (2003)
Kaiser, V. B., Zhou, Q. & Bachtrog, D. Nonrandom gene loss from the Drosophila miranda neo-Y chromosome. Genome Biol. Evol. 3, 1329–1337 (2011)
Jegalian, K. & Page, D. C. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature 394, 776–780 (1998)
Ohno, S. Sex Chromosomes and Sex-linked Genes (Springer-Verlag, 1967)
Huang, N., Lee, I., Marcotte, E. M. & Hurles, M. E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 6, e1001154 (2010)
Carrel, L. & Willard, H. F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404 (2005)
Yang, F., Babak, T., Shendure, J. & Disteche, C. M. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 20, 614–622 (2010)
Merkin, J., Russell, C., Chen, P. & Burge, C. B. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338, 1593–1599 (2012)
Xie, D. et al. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 20, 804–815 (2010)
Flicek, P. et al. Ensembl 2014. Nucleic Acids Res. 42, D749–D755 (2014)
Wang, X., Douglas, K. C., Vandeberg, J. L., Clark, A. G. & Samollow, P. B. Chromosome-wide profiling of X-chromosome inactivation and epigenetic states in fetal brain and placenta of the opossum, Monodelphis domestica. Genome Res. 24, 70–83 (2014)
Cockwell, A., MacKenzie, M., Youings, S. & Jacobs, P. A cytogenetic and molecular study of a series of 45,X fetuses and their parents. J. Med. Genet. 28, 151–155 (1991)
Hook, E. B. & Warburton, D. The distribution of chromosomal genotypes associated with Turner’s syndrome: livebirth prevalence rates and evidence for diminished fetal mortality and severity in genotypes associated with structural X abnormalities or mosaicism. Hum. Genet. 64, 24–27 (1983)
Hassold, T., Benham, F. & Leppert, M. Cytogenetic and molecular analysis of sex-chromosome monosomy. Am. J. Hum. Genet. 42, 534–541 (1988)
Burgoyne, P. S., Tam, P. P. & Evans, E. P. Retarded development of XO conceptuses during early pregnancy in the mouse. J. Reprod. Fertil. 68, 387–393 (1983)
Burgoyne, P. S. & Baker, T. G. Oocyte depletion in XO mice and their XX sibs from 12 to 200 days post partum. J. Reprod. Fertil. 61, 207–212 (1981)
Burgoyne, P. S., Evans, E. P. & Holland, K. XO monosomy is associated with reduced birthweight and lowered weight gain in the mouse. J. Reprod. Fertil. 68, 381–385 (1983)
Lindgren, A. M. et al. Haploinsufficiency of KDM6A is associated with severe psychomotor retardation, global growth restriction, seizures and cleft palate. Hum. Genet. 132, 537–552 (2013)
Rujirabanjerd, S. et al. Identification and characterization of two novel JARID1C mutations: suggestion of an emerging genotype-phenotype correlation. Eur. J. Hum. Genet. 18, 330–335 (2010)
Lawson-Yuen, A., Saldivar, J. S., Sommer, S. & Picker, J. Familial deletion within NLGN4 associated with autism and Tourette syndrome. Eur. J. Hum. Genet. 16, 614–618 (2008)
Huang, L. et al. A noncoding, regulatory mutation implicates HCFC1 in nonsyndromic intellectual disability. Am. J. Hum. Genet. 91, 694–702 (2012)
Ramocki, M. B., Tavyev, Y. J. & Peters, S. U. The MECP2 duplication syndrome. Am. J. Med. Genet. A. 152A, 1079–1088 (2010)
Froyen, G. et al. Copy-number gains of HUWE1 due to replication- and recombination-based rearrangements. Am. J. Hum. Genet. 91, 252–264 (2012)
Lau, E. C., Mohandas, T. K., Shapiro, L. J., Slavkin, H. C. & Snead, M. L. Human and mouse amelogenin gene loci are on the sex chromosomes. Genomics 4, 162–168 (1989)
Rozen, S., Marszalek, J. D., Alagappan, R. K., Skaletsky, H. & Page, D. C. Remarkably little variation in proteins encoded by the Y chromosome’s single-copy genes, implying effective purifying selection. Am. J. Hum. Genet. 85, 923–928 (2009)
Watanabe, M., Zinn, A. R., Page, D. C. & Nishimoto, T. Functional equivalence of human X- and Y-encoded isoforms of ribosomal protein S4 consistent with a role in Turner syndrome. Nature Genet. 4, 268–271 (1993)
Sekiguchi, T., Iida, H., Fukumura, J. & Nishimoto, T. Human DDX3Y, the Y-encoded isoform of RNA helicase DDX3, rescues a hamster temperature-sensitive ET24 mutant cell line with a DDX3X mutation. Exp. Cell Res. 300, 213–222 (2004)
Welstead, G. G. et al. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc. Natl Acad. Sci. USA 109, 13004–13009 (2012)
Shpargel, K. B., Sengoku, T., Yokoyama, S. & Magnuson, T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 8, e1002964 (2012)
Lee, S., Lee, J. W. & Lee, S. K. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev. Cell 22, 25–37 (2012)
Slonim, D., Kruglyak, L., Stein, L. & Lander, E. Building human genome maps with radiation hybrids. J. Comput. Biol. 4, 487–504 (1997)
Saxena, R. et al. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nature Genet. 14, 292–299 (1996)
Yang, Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997)
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004)
Felsenstein, J. PHYLIP - phylogeny inference package (version 3.2). Cladistics 5, 164–166 (1989)
Thomas, P. D. et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003)
The UniProt Consortium Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 41, D43–D47 (2013)
Kuroda-Kawaguchi, T. et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nature Genet. 29, 279–286 (2001)
Karere, G. M., Froenicke, L., Millon, L., Womack, J. E. & Lyons, L. A. A high-resolution radiation hybrid map of rhesus macaque chromosome 5 identifies rearrangements in the genome assembly. Genomics 92, 210–218 (2008)
Mi, H., Muruganujan, A., Casagrande, J. T. & Thomas, P. D. Large-scale gene function analysis with the PANTHER classification system. Nature Protocols 8, 1551–1566 (2013)
Gubbay, J. et al. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 346, 245–250 (1990)
Sinclair, A. H. et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346, 240–244 (1990)
Hayashida, H., Kuma, K. & Miyata, T. Interchromosomal gene conversion as a possible mechanism for explaining divergence patterns of ZFY-related genes. J. Mol. Evol. 35, 181–183 (1992)
Marais, G. & Galtier, N. Sex chromosomes: how X-Y recombination stops. Curr. Biol. 13, R641–R643 (2003)
Iwase, M. et al. The amelogenin loci span an ancient pseudoautosomal boundary in diverse mammalian species. Proc. Natl Acad. Sci. USA 100, 5258–5263 (2003)
Dal Zotto, L. et al. The mouse Mid1 gene: implications for the pathogenesis of Opitz syndrome and the evolution of the mammalian pseudoautosomal region. Hum. Mol. Genet. 7, 489–499 (1998)
Bachtrog, D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics 179, 1513–1525 (2008)
Jobling, M. A. et al. Structural variation on the short arm of the human Y chromosome: recurrent multigene deletions encompassing Amelogenin Y. Hum. Mol. Genet. 16, 307–316 (2007)
Jones, M. H. et al. The Drosophila developmental gene fat facets has a human homologue in Xp11.4 which escapes X-inactivation and has related sequences on Yq11.2. Hum. Mol. Genet. 5, 1695–1701 (1996)
Adachi, M., Tachibana, K., Asakura, Y., Muroya, K. & Del Ogata, T. Del(X)(p21.1) in a mother and two daughters: genotype-phenotype correlation of Turner features. Hum. Genet. 106, 306–310 (2000)
Chocholska, S., Rossier, E., Barbi, G. & Kehrer-Sawatzki, H. Molecular cytogenetic analysis of a familial interstitial deletion Xp22.2-22.3 with a highly variable phenotype in female carriers. Am. J. Med. Genet. A. 140A, 604–610 (2006)
Good, C. D. et al. Dosage-sensitive X-linked locus influences the development of amygdala and orbitofrontal cortex, and fear recognition in humans. Brain 126, 2431–2446 (2003)
James, R. S. et al. A study of females with deletions of the short arm of the X chromosome. Hum. Genet. 102, 507–516 (1998)
Massa, G., Vanderschueren-Lodeweyckx, M. & Fryns, J. P. Deletion of the short arm of the X chromosome: a hereditary form of Turner syndrome. Eur. J. Pediatr. 151, 893–894 (1992)
Zinn, A. R. et al. Del (X)(p21.2) in a mother and two daughters with variable ovarian function. Clin. Genet. 52, 235–239 (1997)
Zinn, A. R. et al. Evidence for a Turner syndrome locus or loci at Xp11.2-p22.1. Am. J. Hum. Genet. 63, 1757–1766 (1998)
Acknowledgements
We thank W. J. Murphy, E. Owens and J. E. Womak for generating radiation hybrid panels and for assistance in mapping; L. Lyons and W.J.M. for providing the rhesus radiation hybrid panel; A. Kaur for a rhesus cell line; S. Austad, P. Hornsby and S. Tardif for marmoset cell lines; M. Brown for rat cell lines; J.E.W. for bull fibroblasts; W. Johnson and S. O’Neil for rhesus tissues samples; W.J., S.O. and S.T. for marmoset tissue samples; M. Turner for rat tissue samples; J.E.W. for bull tissue samples; P. Samollow for opossum tissue samples; E. Vallender for Tamarin DNA; B. Chowdhary and T. Raudsepp for FISH experiments in the bull; C. Friedman and B. Trask for flow-sorted marmoset Y chromosomes; B.T. for sizing rat Y chromosomes; C. Burge for permission to assemble transcriptome data from SRR594455, SRR594463 and SRR594508; J. Alföldi for permission to assemble transcriptome data from SRR500909; R.B. Norgren for permission to assemble transcriptome data from SRR544870; and A. Godfrey, Y. Hu and B. Lesch for comments on the manuscript. Supported by National Institutes of Health and Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
D.W.B., J.F.H., H.S., S. Rozen, W.C.W., R.A.G., R.K.W. and D.C.P. planned the project. J.F.H., H.S., L.G.B., T.-J.C., N.K., S.Z. and J.A. performed BAC mapping, radiation hybrid mapping and real-time polymerase chain reaction analyses. T.G., S. Rock, C.K., R.S.F., S.D., Y.D., D.M., Z.K., L.L., C.B., Q.W., J.W., M.H., S.L., L.N. and D.M.M. were responsible for BAC sequencing. D.W.B., J.F.H. and H.S. performed comparative sequence analyses. T.P. performed FISH analyses. D.W.B. and D.C.P. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Annotated sequence contigs from the MSY of five species.
All sequence features and BACs drawn to scale. a–e, Marmoset MSY. f–j, Mouse MSY. k–o, Rat MSY. p–t, Bull MSY. u–y, Opossum MSY. a, f, k, p, u, Schematic representation of assembled contigs and sequence classes: X-degenerate (yellow); ampliconic (blue); pseudoautosomal (green); heterochromatic (pink). Gaps shown in white. b, g, l, q, v, Positions of all intact, actively transcribed genes. Plus (+) strand above, minus (−) strand below. c, h, m, r, w, G + C content (%) calculated in a 100-kb sliding window with 1-kb steps. d, i, n, s, x, Alu (red), LINE (green), and endogenous retrovirus (blue) content (%) calculated in a 200-kb sliding window with 1-kb steps. e, j, o, t, y, Sequenced MSY BACs. Each bar represents the size and position of one BAC clone, labelled with the library identifier. e, BAC clones with no prefix are from the CHORI-259 library; BAC clones with “A” prefix are from the MARMAEX (Amplicon Express) library. j, All BAC clones are from the RPCI-24 library. o, BAC clones without prefix are from the RNAEX library; BAC clones with “E” prefix are from the RNECO library (both from Amplicon Express). t, BAC clones without prefix are from the CHORI-240 library; BAC clones with “E” prefix are from the BTDAEX library (both from Amplicon Express). y, BAC clones with no prefix are from the VMRC6 library; BAC clones with “A” prefix are from the MDAEX (Amplicon Express) library.
Extended Data Figure 2 Expression of Y-linked genes across tissues and species.
Within each species, the relative expression of each Y-linked gene is shown as a heat map normalized to the male tissue with the highest level of expression of that gene. Expression was calculated from RNA-seq data as reads per kilobase of transcript per million mapped reads. For each gene and species, tissues are arranged in alphabetical order from left to right: brain, cerebellum, colon, heart, kidney, liver, lung, skeletal muscle, spleen and testis. Most single-copy genes (red) are broadly expressed across male tissues, whereas Y-linked genes in multi-copy families (blue) are predominantly or exclusively expressed in testes.
Extended Data Figure 3 Dot plot of human X orthologues in opossum and chicken.
Rectangular dot plots show chromosomal locations of X-orthologous genes in other species. The human X chromosome is composed of a conserved region, orthologous to the opossum X chromosome and a region of chicken chromosome 4, as well as an added region, orthologous to chicken chromosome 1, which has broken in two in the opossum lineage.
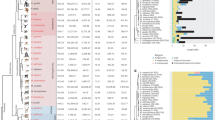
Extended Data Figure 4 Phylogenetic analysis of stratum one and stratum two genes.
Consensus phylogenies reconstructed by DNAML with 100 bootstrap replicates; scale bars represent the expected number of nucleotide substitutions per site along each branch. Phylogenies for ancestral X–Y pair genes from the X-conserved region, shared between placental and marsupial mammals are shown. Adjacent to each tree, pink and light blue bars highlight the positions of the X and Y homologues, respectively; red and dark blue bars highlight the position of placental and marsupial homologues, respectively. Among the three gene pairs from stratum one (SOX3/SRY, RBMX/RBMY, and HSFX/HSFY), Y-linked genes are more closely related to each other than their X-linked orthologues. Among the other gene pairs (KDM5C/KDM5D and UBE1X/UBE1Y), marsupial X–Y pairs are more closely related to each other than they are to placental orthologues, suggesting that a second stratum formed independently in the placental and marsupial ineages. Species abreviations: HAS, human; PTR, chimpanzee; MAQ, rhesus; CJA, marmoset; MUS, mouse; RNO, rat; BTA, bull; MDO, opossum; and GGA, chicken.
Extended Data Figure 5 Phylogenetic tree showing X–Y gene conversion in AMELX/AMELY and ZFX/ZFY.
Consensus phylogenies reconstructed by DNAML with 100 bootstrap replicates; scale bars represent the expected number of nucleotide substitutions per site along each branch. Phylogenies of three ancestral X–Y pair genes from the placental-specific X-added region within stratum 2/3 (USP9X/USP9Y, AMELX/AMELY and ZFX/ZFY) are shown. Within each tree, pink and light blue branches highlight the positions of the X and Y homologues, respectively. USP9X/USP9Y is a typical stratum 2/3 gene pair; all USP9Y genes are more closely related to each other than to any USP9X gene. AMELX/AMELY and ZFX/ZFY show more complex histories. For example, bull AMELY is more closely related to bull AMELX than to any other AMELY orthologue. X–Y gene conversion occurred after stratum formation in multiple lineages. Species abreviations: HAS, human; PTR, chimpanzee; MAQ, rhesus; CJA, marmoset; MUS, mouse; RNO, rat; BTA, bull; MDO, opossum; GGA, chicken; and XTR, Xenopus tropicalis.
Extended Data Figure 6 Y–Y gene conversion within multi-copy gene families.
Consensus phylogenies reconstructed by DNAML with 100 bootstrap replicates; scale bars represent the expected number of nucleotide substitutions per site along each branch. Phylogenies for ancestral X–Y pair genes from the X-conserved region, shared between placental and marsupial mammals are shown. Adjacent to each tree, light blue bars highlight the positions of Y-linked genes with high within-species identity and across-species divergence, indicating that gene conversion is more frequent than mutation. a-g, TSPY, RBMY, SRY, HSFY, DDX3Y, UBE1Y and EIF1AY show signs of Y–Y gene conversion; in the species where they are present in multiple copies, they are clustered in arrays of genes. h, i, RPS4Y and ZFY do not show signs of recent Y–Y gene conversion; in the species where they are present in two copies, they are dispersed on the Y chromosome. a, TSPY is present as a multi-copy gene family on the human, chimpanzee, rhesus, marmoset and bull Y chromosomes. Note that 2 distinct families of TSPY emerged in bull. b, RBMY is present as a multi-copy gene family on the human, chimpanzee, marmoset, mouse and bull Y chromosomes. c, SRY is present as a multi-copy gene family on the rat Y chromosome. d, HSFY is present as a multi-copy gene family on the human, rhesus, and bull Y chromosomes. e, DDX3Y is present as a multi-copy gene family on the marmoset Y chromosome. f, UBE1Y is present as a multi-copy gene family on the rat Y chromosome. g, EIF1AY is present as a multi-copy gene family on the marmoset Y chromosome. h, RPS4Y is present is present as a multi-copy gene family on the human, chimpanzee and rhesus Y chromosomes. RPS4Y genes appear to have split into two distinct families before the divergence of primate species, which have not engaged in subsequent gene conversion within each species. i, ZFY is present as a multi-copy gene family on the mouse Y chromosome. Although ZFY participated in multiple independent X–Y gene conversion events after the divergence of placental mammals, there is no evidence of recent Y–Y gene conversion in mouse. Mouse Zfy1 and Zfy2 genes are more divergent than human and chimpanzee ZFY. Species abbreviations: HAS, human; PTR, chimpanzee; MAQ, rhesus; CJA, marmoset; MUS, mouse; RNO, rat; BTA, bull; MDO, opossum; GGA, chicken; MFA, Macaca fascicularis; and XTR, Xenopus tropicalis.
Extended Data Figure 7 Viable structually variant sex chromosomes in humans.
The presence of the 12 broadly expressed, dosage-sensitive X–Y pair genes and other chromosomal features on structurally variant sex chromosomes are indicated by filled circles. a, Viable non-mosaic deletions of X–Y pair genes from the human Y chromosome. The human Y chromosome is susceptible to structural rearrangements due to homology mediated crossing-over between repeated sequences. Crossing-over between tandem repeats creates interstitial deletions, whereas crossing-over in palindrome arms causes the formation of isodicentric chromosomes and isochromosomes. Each Y-linked member of the 12, broadly expressed, dosage-sensitive X–Y gene pairs is deleted in one or more variants, thus no single X–Y pair gene is haplolethal. b, Viable deletions of X–Y pair genes from the human X chromosome in females are shown. Reported cases of X chromosome deletions in females are consistent with a collective haplolethality for all 12 broadly expressed, dosage-sensitive X–Y gene pairs in humans. Familial cases, where a variant X chromosome has been transmitted from mother to daughter, are unlikely to be mosaic. The most extensive deletion among familial cases eliminates 7 of 12 genes. The most extensive de novo deletion variants eliminate 11 of 12 genes, but mosaicism for 46,XX cells cannot be excluded. No variants remove RPS4X because of viability effects mediated by its position between the centromere (CEN) and X-inactivation centre (XIC) on the long arm, rather than haplolethality of RPS4X alone.
Supplementary information
Supplementary Data 1
This data file contains FASTA alignment of X-Y pairs with GGA ORFs. (TXT 1246 kb)
Supplementary Data 2
This data file contains FASTA sequences used to generate alignments. (TXT 860 kb)
Supplementary Tables
This file contains Supplementary Tables 1-5. (XLSX 371 kb)
Rights and permissions
About this article
Cite this article
Bellott, D., Hughes, J., Skaletsky, H. et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508, 494–499 (2014). https://doi.org/10.1038/nature13206
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13206
This article is cited by
-
Muscle miRNAs are influenced by sex at baseline and in response to exercise
BMC Biology (2023)
-
Sexual dimorphism in chronic respiratory diseases
Cell & Bioscience (2023)
-
Developmental transcriptomic patterns can be altered by transgenic overexpression of Uty
Scientific Reports (2023)
-
Trio-binning of a hinny refines the comparative organization of the horse and donkey X chromosomes and reveals novel species-specific features
Scientific Reports (2023)
-
Pericentromeric recombination suppression and the ‘large X effect’ in plants
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.