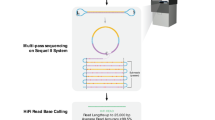
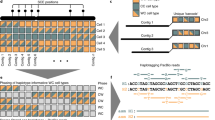
Abstract
The reference sequences of structurally complex regions can be obtained only through highly accurate clone-based approaches. We and others have successfully used single-haplotype iterative mapping and sequencing (SHIMS) 1.0 to assemble structurally complex regions across the sex chromosomes of several vertebrate species and to allow for targeted improvements to the reference sequences of human autosomes. However, SHIMS 1.0 is expensive and time consuming, requiring resources that only a genome center can provide. Here we introduce SHIMS 2.0, an improved SHIMS protocol that allows even a small laboratory to generate high-quality reference sequence from complex genomic regions. Using a streamlined and parallelized library-preparation protocol, and taking advantage of inexpensive high-throughput short-read-sequencing technologies, a small laboratory with both molecular biology and bioinformatics experience can sequence and assemble 192 large-insert bacterial artificial chromosome (BAC) or fosmid clones in 1 week. In SHIMS 2.0, in contrast to other pooling strategies, each clone is sequenced with a unique barcode, thus enabling clones containing nearly identical sequences to be multiplexed in a single sequencing run and assembled separately. Relative to SHIMS 1.0, SHIMS 2.0 decreases the required cost and time by two orders of magnitude while preserving high sequencing accuracy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Mueller, J.L. et al. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat. Genet. 45, 1083–1087 (2013).
Lupski, J.R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 14, 417–422 (1998).
Stankiewicz, P. & Lupski, J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61, 437–455 (2010).
Ross, M.T. et al. The DNA sequence of the human X chromosome. Nature 434, 325–337 (2005).
International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004).
Gordon, D. & Green, P. Consed: a graphical editor for next-generation sequencing. Bioinformatics 29, 2936–2937 (2013).
Bonfield, J.K., Smith, K. & Staden, R. A new DNA sequence assembly program. Nucleic Acids Res. 23, 4992–4999 (1995).
She, X. et al. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature 431, 927–930 (2004).
Alkan, C., Sajjadian, S. & Eichler, E.E. Limitations of next-generation genome sequence assembly. Nat. Methods 8, 61–65 (2011).
Gordon, D. et al. Long-read sequence assembly of the gorilla genome. Science 352, aae0344 (2016).
Eichler, E.E. Segmental duplications: what's missing, misassigned, and misassembled—and should we care? Genome Res. 11, 653–656 (2001).
Dennis, M.Y. et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 149, 912–922 (2012).
Steinberg, K.M. et al. Single haplotype assembly of the human genome from a hydatidiform mole. Genome Res. 24, 2066–2076 (2014).
Watson, C.T. et al. Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am. J. Hum. Genet. 92, 530–546 (2013).
Mohajeri, K. et al. Interchromosomal core duplicons drive both evolutionary instability and disease susceptibility of the chromosome 8p23.1 region. Genome Res. 26, 1453–1467 (2016).
Sachidanandam, R. et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409, 928–933 (2001).
Kuroda-Kawaguchi, T. et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat. Genet. 29, 279–286 (2001).
Skaletsky, H. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003).
Repping, S. et al. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat. Genet. 38, 463–467 (2006).
Lange, J. et al. Intrachromosomal homologous recombination between inverted amplicons on opposing Y-chromosome arms. Genomics 102, 257–264 (2013).
Lange, J., Skaletsky, H., Bell, G.W. & Page, D.C. MSY Breakpoint Mapper, a database of sequence-tagged sites useful in defining naturally occurring deletions in the human Y chromosome. Nucleic Acids Res. 36, D809 D (2008).
Lange, J. et al. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell 138, 855–869 (2009).
Repping, S. et al. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat. Genet. 35, 247–251 (2003).
Repping, S. et al. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am. J. Hum. Genet. 71, 906–922 (2002).
Repping, S. et al. A family of human Y chromosomes has dispersed throughout northern Eurasia despite a 1.8-Mb deletion in the azoospermia factor c region. Genomics 83, 1046–1052 (2004).
Rozen, S.G. et al. AZFc deletions and spermatogenic failure: a population-based survey of 20,000 Y chromosomes. Am. J. Hum. Genet. 91, 890–896 (2012).
Bellott, D.W. et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508, 494–499 (2014).
Bellott, D.W. et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466, 612–616 (2010).
Hughes, J.F. et al. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 483, 82–86 (2012).
Hughes, J.F. et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 463, 536–539 (2010).
Soh, Y.Q. et al. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 159, 800–813 (2014).
Bellott, D.W. et al. Avian W and mammalian Y chromosomes convergently retained dosage-sensitive regulators. Nat. Genet. 49, 387–394 (2017).
Li, G. et al. Comparative analysis of mammalian Y chromosomes illuminates ancestral structure and lineage-specific evolution. Genome Res. 23, 1486–1495 (2013).
Sato, K., Motoi, Y., Yamaji, N. & Yoshida, H. 454 Sequencing of pooled BAC clones on chromosome 3H of barley. BMC Genom. 12, 246 (2011).
Quinn, N.L. et al. Assessing the feasibility of GS FLX Pyrosequencing for sequencing the Atlantic salmon genome. BMC Genom. 9, 404 (2008).
Rounsley, S., Lin, X. & Ketchum, K.A. Large-scale sequencing of plant genomes. Curr. Opin. Plant Biol. 1, 136–141 (1998).
National Center for Biotechnology Information. Commercial and Academic Suppliers of Clones, Libraries and Other Reagents Described in Clone DB https://www.ncbi.nlm.nih.gov/clone/content/distributors/ (2017).
Guha, S. & Maheshwari, S.C. Cell division and differentiation of embryos in pollen grains of Daturain vitro. Nature 212, 97–98 (1966).
Jain, S.M., Sopory, S.K. & Veilleux, R.E. In vitro haploid production in higher plants (Kluwer Academic Publishers, 1996).
Bonfield, J.K. & Whitwham, A. Gap5: editing the billion fragment sequence assembly. Bioinformatics 26, 1699–1703 (2010).
Rohland, N. & Reich, D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22, 939–946 (2012).
Wilkening, S. et al. Genotyping 1000 yeast strains by next-generation sequencing. BMC Genom. 14, 90 (2013).
Gymrek, M., Golan, D., Rosset, S. & Erlich, Y. lobSTR: a short tandem repeat profiler for personal genomes. Genome Res. 22, 1154–1162 (2012).
Goodwin, S. et al. Oxford nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 25, 1750–1756 (2015).
Berlin, K. et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33, 623–630 (2015).
Koren, S. et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol. 30, 693–700 (2012).
Madoui, M.A. et al. Genome assembly using nanopore-guided long and error-free DNA reads. BMC Genom. 16, 327 (2015).
Tomaszkiewicz, M. et al. A time- and cost-effective strategy to sequence mammalian Y chromosomes: an application to the de novo assembly of gorilla Y. Genome Res. 26, 530–540 (2016).
McCoy, R.C. et al. Illumina TruSeq synthetic long-reads empower de novo assembly and resolve complex, highly-repetitive transposable elements. PLoS One 9, e106689 (2014).
Li, R. et al. Illumina synthetic long read sequencing allows recovery of missing sequences even in the “finished” C. elegans genome. Sci. Rep. 5, 10814 (2015).
Dong, Y. et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol. 31, 135–141 (2013).
Seo, J.S. et al. De novo assembly and phasing of a Korean human genome. Nature 538, 243–247 (2016).
Nagaraja, R. et al. Characterization of four human YAC libraries for clone size, chimerism and X chromosome sequence representation. Nucleic Acids Res. 22, 3406–3411 (1994).
Venter, J.C., Smith, H.O. & Hood, L. A new strategy for genome sequencing. Nature 381, 364–366 (1996).
Glenn, T.C. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 11, 759–769 (2011).
Agencourt Bioscience Corporation. Agencourt CosMCPrep High and Low Copy Plasmid Purification https://www.beckmancoulter.com/wsrportal/bibliography?docname=Protocol000381v012.pdf (2006).
Lange, V. et al. Cost-efficient high-throughput HLA typing by MiSeq amplicon sequencing. BMC Genom. 15, 63 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Sahlin, K., Vezzi, F., Nystedt, B., Lundeberg, J. & Arvestad, L. BESST: efficient scaffolding of large fragmented assemblies. BMC Bioinform. 15, 281 (2014).
Salmela, L., Sahlin, K., Makinen, V. & Tomescu, A.I. Gap filling as exact path length problem. J. Comput. Biol. 23, 347–361 (2016).
Church, D.M. Tiling Path File (TPF) Specification v1.4 https://www.ncbi.nlm.nih.gov/projects/genome/assembly/TPF_Specification_v1.4_20110215.pdf (2011).
National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/assembly/agp/AGP_Specification/ (2014).
McPherson, J.D. et al. A physical map of the human genome. Nature 409, 934–941 (2001).
National Center for Biotechnology Information. What is tbl2asn? https://www.ncbi.nlm.nih.gov/genbank/tbl2asn2/ (2017).
Acknowledgements
This work was supported by the National Institutes of Health and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
D.W.B., H.S., J.F.H., and D.C.P. designed the study. D.W.B. and T.-J.C. developed the experimental methods. D.W.B. wrote the scripts for computational analysis. D.W.B., T.-J.C., and D.C.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Table 1 (PDF 351 kb)
Rights and permissions
About this article
Cite this article
Bellott, D., Cho, TJ., Hughes, J. et al. Cost-effective high-throughput single-haplotype iterative mapping and sequencing for complex genomic structures. Nat Protoc 13, 787–809 (2018). https://doi.org/10.1038/nprot.2018.019
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2018.019
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.