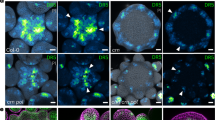
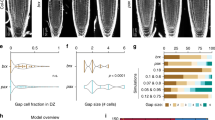
Abstract
How biological systems generate reproducible patterns with high precision is a central question in science1. The shoot apical meristem (SAM), a specialized tissue producing plant aerial organs, is a developmental system of choice to address this question. Organs are periodically initiated at the SAM at specific spatial positions and this spatiotemporal pattern defines phyllotaxis. Accumulation of the plant hormone auxin triggers organ initiation2,3,4,5, whereas auxin depletion around organs generates inhibitory fields that are thought to be sufficient to maintain these patterns and their dynamics4,6,7,8,9,10,11,12,13. Here we show that another type of hormone-based inhibitory fields, generated directly downstream of auxin by intercellular movement of the cytokinin signalling inhibitor ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (AHP6)14, is involved in regulating phyllotactic patterns. We demonstrate that AHP6-based fields establish patterns of cytokinin signalling in the meristem that contribute to the robustness of phyllotaxis by imposing a temporal sequence on organ initiation. Our findings indicate that not one but two distinct hormone-based fields may be required for achieving temporal precision during formation of reiterative structures at the SAM, thus indicating an original mechanism for providing robustness to a dynamic developmental system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lander, A. D. Pattern, growth, and control. Cell 144, 955–969 (2011)
Vernoux, T., Kronenberger, J., Grandjean, O., Laufs, P. & Traas, J. PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127, 5157–5165 (2000)
Reinhardt, D., Mandel, T. & Kuhlemeier, C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12, 507–518 (2000)
Reinhardt, D. et al. Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260 (2003)
Heisler, M. G. et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899–1911 (2005)
Mitchison, G. J. Phyllotaxis and the Fibonacci series. Science 196, 270–275 (1977)
Veen, A. H. & Lindenmayer, A. Diffusion mechanism for phyllotaxis: theoretical physico-chemical and computer study. Plant Physiol. 60, 127–139 (1977)
Douady, S. & Couder, Y. Phyllotaxis as a dynamical self organizing process. Part II: the spontaneous formation of a periodicity and the coexistence of spiral and whorled patterns. J. Theor. Biol. 178, 275–294 (1996)
de Reuille, P. B. et al. Computer simulations reveal properties of the cell–cell signaling network at the shoot apex in Arabidopsis. Proc. Natl Acad. Sci. USA 103, 1627–1632 (2006)
Jönsson, H., Heisler, M. G., Shapiro, B. E., Meyerowitz, E. M. & Mjolsness, E. An auxin-driven polarized transport model for phyllotaxis. Proc. Natl Acad. Sci. USA 103, 1633–1638 (2006)
Smith, R. S. et al. A plausible model of phyllotaxis. Proc. Natl Acad. Sci. USA 103, 1301–1306 (2006)
Stoma, S. et al. Flux-based transport enhancement as a plausible unifying mechanism for auxin transport in meristem development. PLOS Comput. Biol. 4, e1000207 (2008)
Vernoux, T. et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7, 508 (2011)
Mähönen, A. P. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311, 94–98 (2006)
Giulini, A., Wang, J. & Jackson, D. Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430, 1031–1034 (2004)
Leibfried, A. et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175 (2005)
Yadav, R. K., Girke, T., Pasala, S., Xie, M. & Reddy, G. V. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl Acad. Sci. USA 106, 4941–4946 (2009)
Bartrina, I., Otto, E., Strnad, M., Werner, T. & Schmülling, T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23, 69–80 (2011)
Couder, Y. Initial transitions, order and disorder in phyllotactic patterns: the ontogeny of Helanthus annuus. A case study. Acta Societates Botanicarum Poloniae 67, 129–150 (1998)
Benková, E. et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 (2003)
Laufs, P., Grandjean, O., Jonak, C., Kiêu, K. & Traas, J. Cellular parameters of the shoot apical meristem in Arabidopsis. Plant Cell 10, 1375–1390 (1998)
Bishopp, A. et al. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 21, 917–926 (2011)
Hardtke, C. S. & Berleth, T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411 (1998)
To, J. P. C. et al. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16, 658–671 (2004)
Müller, B. & Sheen, J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094–1097 (2008)
Dello Ioio, R. et al. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384 (2008)
Marhavý, P. et al. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev. Cell 21, 796–804 (2011)
Zhao, Z. et al. Hormonal control of the shoot stem-cell niche. Nature 465, 1089–1092 (2010)
Mirabet, V., Besnard, F., Vernoux, T. & Boudaoud, A. Noise and robustness in phyllotaxis. PLoS Comput. Biol. 8, e1002389 (2012)
Fernandez, R. et al. Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nature Methods 7, 547–553 (2010)
Deveaux, Y. et al. The ethanol switch: a tool for tissue-specific gene induction during plant development. Plant J. 36, 918–930 (2003)
Yanai, O. et al. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15, 1566–1571 (2005)
Schlereth, A. et al. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464, 913–916 (2010)
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998)
Lee, J.-Y. et al. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc. Natl Acad. Sci. USA 103, 6055–6060 (2006)
Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S. & Mullineaux, P. M. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832 (2000)
Karimi, M., De Meyer, B. & Hilson, P. Modular cloning in plant cells. Trends Plant Sci. 10, 103–105 (2005)
Udvardi, M. K., Czechowski, T. & Scheible, W.-R. Eleven golden rules of quantitative RT–PCR. Plant Cell 20, 1736–1737 (2008)
Rieu, I. & Powers, S. J. Real-time quantitative RT–PCR: design, calculations, and statistics. Plant Cell 21, 1031–1033 (2009)
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515 (2004)
Wu, Z., Irizarry, R. A., Gentleman, R., Martinez-Murillo, F. & Spencer, F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99, 909–917 (2004)
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, http://dx.doi.org/10.2202/1544-6115.1027 (2004)
Storey, J. D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA 100, 9440–9445 (2003)
Dümmler, A., Lawrence, A.-M. & de Marco, A. Simplified screening for the detection of soluble fusion constructs expressed in E. coli using a modular set of vectors. Microb. Cell Fact. 4, 34 (2005)
Hejátko, J. et al. In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nature Protocols 1, 1939–1946 (2006)
Grandjean, O. et al. In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell 16, 74–87 (2004)
Hotton, S. Finding the center of a phyllotactic pattern. J. Theor. Biol. 225, 15–32 (2003)
Barbier de Reuille, P. B., Bohn-Courseau, I., Godin, C. & Traas, J. A protocol to analyse cellular dynamics during plant development. Plant J. 44, 1045–1053 (2005)
Stoma, S., Fröhlich, M., Gerber, S. & Klipp, E. STSE: Spatio-temporal simulation environment dedicated to biology. BMC Bioinformatics 12, 126 (2011)
Pradal, C., Boudon, F., Nouguier, C., Chopard, J. & Godin, C. PlantGL: a Python-based geometric library for 3D plant modelling at different scales. Graph. Models 71, 1–21 (2009)
Peaucelle, A., Morin, H., Traas, J. & Laufs, P. Plants expressing a miR164-resistant CUC2 gene reveal the importance of post-meristematic maintenance of phyllotaxy in Arabidopsis. Development 134, 1045–1050 (2007)
Refahi, Y. et al. in Combinatorial Pattern Matching (Lecture Notes in Computer Science) Vol. 6661 323–335 (Springer, 2011)
Guédon, Y. et al. Pattern identification and characterization reveal permutations of organs as a key genetically controlled property of post-meristematic phyllotaxis. J. Theor. Biol. (2013)
Mardia, K. V. & Jupp, P. E. Directional statistics (Wiley, 2000)
Csiszár, I. & Talata, Z. Context tree estimation for not necessarily finite memory processes, via BIC and MDL. IEEE Trans. Inf. Theory 52, 1007–1016 (2006)
Acknowledgements
We thank M. Heisler, B. Müller and D. Weijers for sharing materials; A. Miyawaki for VENUS; D. Mast for help with plant analysis; C. Chamot and C. Lionnet (PLATIM) for help with confocal microscopy and ImageJ (C. Chamot); S. Chamot for help with live imaging; C. Gauthier and A. Laugraud (PRABI, Université Lyon I) for help with statistical analyses; Xavier Jaurand (Pi2) for help with the SEM; Y. Couder, S. Douady, M. Bennett and M.-A. Félix for their insights and support; Y. Jaillais, O. Hamant and C. Scutt for comments on the manuscript. T.V. was supported by HFSPO CDA 0047/2007 (Human Frontier Science Program Organization) and ANR-07-JCJC-0115 (Agence National de la Recherche) grants; R.D., F.P. and T.V. by the ANR-12-BSV6-0005 grant (AuxiFlo); J.L., J.T., C.G., Y.H. and T.V. by a transnational EraSysBio Grant (iSAM); F.B. by a predoctoral grant of the French Ministry of Research; and Y.R. by a CJS grant from INRA.
Author information
Authors and Affiliations
Contributions
F.B. and T.V. conceived and designed the experiments. F.B., V.Mo., B.M., G.B., P.C., F.R., J.L., S.L., C.C., E.T., P.D. and T.V. performed the experiments. Y.R., E.F., C.G., F.B., G.B., T.V. and Y.G. performed the mathematical analysis of phyllotaxis. V.Mi., G.B. and A.Bo. analysed the auxin transport network. R.D., F.P., A.Bi., Y.H. and J.T. provided reagents/materials. F.B. and T.V. analysed the data with inputs from all the authors. F.B. and T.V. wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-15 and Supplementary Tables 1-3. (PDF 7993 kb)
Live-imaging of DR5::VENUS and pAHP6::GFP expression
The expression of DR5::VENUS (Magenta) and pAHP6::GFP (Green) was followed over time in the shoot apical meristem using confocal microscopy. (MOV 158 kb)
Rights and permissions
About this article
Cite this article
Besnard, F., Refahi, Y., Morin, V. et al. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 505, 417–421 (2014). https://doi.org/10.1038/nature12791
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12791
This article is cited by
-
Multi-Knock—a multi-targeted genome-scale CRISPR toolbox to overcome functional redundancy in plants
Nature Plants (2023)
-
Phytohormones: plant switchers in developmental and growth stages in potato
Journal of Genetic Engineering and Biotechnology (2021)
-
What shoots can teach about theories of plant form
Nature Plants (2021)
-
The diverse roles of cytokinins in regulating leaf development
Horticulture Research (2021)
-
Developmental stochasticity and variation in floral phyllotaxis
Journal of Plant Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.