Abstract
Plant body plans are elaborated in response to both environmental and endogenous cues. How these inputs intersect to promote growth and development remains poorly understood. During reproductive development, central zone stem cell proliferation in inflorescence meristems is negatively regulated by the CLAVATA3 (CLV3) peptide signalling pathway. In contrast, floral primordia formation on meristem flanks requires the hormone auxin. Here we show that CLV3 signalling is also necessary for auxin-dependent floral primordia generation and that this function is partially masked by both inflorescence fasciation and heat-induced auxin biosynthesis. Stem cell regulation by CLAVATA signalling is separable from primordia formation but is also sensitized to temperature and auxin levels. In addition, we uncover a novel role for the CLV3 receptor CLAVATA1 in auxin-dependent meristem maintenance in cooler environments. As such, CLV3 signalling buffers multiple auxin-dependent shoot processes across divergent thermal environments, with opposing effects on cell proliferation in different meristem regions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Identifiers for published or publicly available lines are provided in Methods. RNA-seq data have been deposited in the NCBI SRA database under BioProject PRJNA661065. All other relevant data are available from the corresponding author upon request.
Code availability
All code used to analyse the data is published in the Nimchuk Lab Github (https://github.com/NimchukLab).
References
de Jong, M. & Leyser, O. Developmental plasticity in plants. Cold Spring Harb. Symp. Quant. Biol. 77, 63–73 (2012).
Weigel, D. Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol. 158, 2–22 (2012).
Chen, Z., Galli, M. & Gallavotti, A. Mechanisms of temperature-regulated growth and thermotolerance in crop species. Curr. Opin. Plant Biol. 65, 102134 (2022).
Reinhardt, D., Mandel, T. & Kuhlemeier, C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12, 507–518 (2000).
Benkova, E. et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 (2003).
Smyth, D. R., Bowman, J. L. & Meyerowitz, E. M. Early flower development in Arabidopsis. Plant Cell 2, 755–767 (1990).
Pernisova, M. & Vernoux, T. Auxin does the SAMba: auxin signaling in the shoot apical meristem. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a039925 (2021).
Willoughby, A. C. & Nimchuk, Z. L. WOX going on: CLE peptides in plant development. Curr. Opin. Plant Biol. 63, 102056 (2021).
Fletcher, J. C. Recent advances in Arabidopsis CLE peptide signaling. Trends Plant Sci. 25, 1005–1016 (2020).
Xu, C. et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47, 784–792 (2015).
Liu, L. et al. Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat. Plants 7, 287–294 (2021).
Clark, S. E., Williams, R. W. & Meyerowitz, E. M. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585 (1997).
Jeong, S., Trotochaud, A. E. & Clark, S. E. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11, 1925–1934 (1999).
Muller, R., Bleckmann, A. & Simon, R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20, 934–946 (2008).
Nimchuk, Z. L., Tarr, P. T. & Meyerowitz, E. M. An evolutionarily conserved pseudokinase mediates stem cell production in plants. Plant Cell 23, 851–854 (2011).
Bleckmann, A., Weidtkamp-Peters, S., Seidel, C. A. & Simon, R. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152, 166–176 (2010).
Schoof, H. et al. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644 (2000).
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M. & Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619 (2000).
Mayer, K. F. et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815 (1998).
DeYoung, B. J. et al. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45, 1–16 (2006).
Nimchuk, Z. L., Zhou, Y., Tarr, P. T., Peterson, B. A. & Meyerowitz, E. M. Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142, 1043–1049 (2015).
Nimchuk, Z. L. CLAVATA1 controls distinct signaling outputs that buffer shoot stem cell proliferation through a two-step transcriptional compensation loop. PLoS Genet. 13, e1006681 (2017).
Rodriguez-Leal, D. et al. Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat. Genet. 51, 786–792 (2019).
Nimchuk, Z. L., Tarr, P. T., Ohno, C., Qu, X. & Meyerowitz, E. M. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr. Biol. 21, 345–352 (2011).
Crook, A. D. et al. BAM1/2 receptor kinase signaling drives CLE peptide-mediated formative cell divisions in Arabidopsis roots.Proc. Natl Acad. Sci. USA 117, 32750–32756 (2020).
Ogawa, M., Shinohara, H., Sakagami, Y. & Matsubayashi, Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294 (2008).
Roman, A. O. et al. HSL1 and BAM1/2 impact epidermal cell development by sensing distinct signaling peptides. Nat. Commun. 13, 876 (2022).
Anne, P. et al. CLERK is a novel receptor kinase required for sensing of root-active CLE peptides in Arabidopsis. Development https://doi.org/10.1242/dev.162354 (2018).
Shinohara, H., Moriyama, Y., Ohyama, K. & Matsubayashi, Y. Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. Plant J. 70, 845–854 (2012).
Shinohara, H. & Matsubayashi, Y. Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. Plant J. 82, 328–336 (2015).
Jones, D. S., John, A., VanDerMolen, K. R. & Nimchuk, Z. L. CLAVATA signaling ensures reproductive development in plants across thermal environments. Curr. Biol. 31, 220–227.e5 (2021).
Schlegel, J. et al. Control of Arabidopsis shoot stem cell homeostasis by two antagonistic CLE peptide signalling pathways. eLife https://doi.org/10.7554/eLife.70934 (2021).
Hord, C. L., Chen, C., Deyoung, B. J., Clark, S. E. & Ma, H. The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18, 1667–1680 (2006).
Cui, Y. et al. CIK receptor kinases determine cell fate specification during early anther development in Arabidopsis. Plant Cell 30, 2383–2401 (2018).
Zhu, Y. et al. Conserved and differentiated functions of CIK receptor kinases in modulating stem cell signaling in Arabidopsis. Mol. Plant 14, 1119–1134 (2021).
Hu, C. et al. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants 4, 205–211 (2018).
Hu, C. et al. A CLE-BAM-CIK signalling module controls root protophloem differentiation in Arabidopsis. New Phytol. 233, 282–296 (2022).
Zhao, Y. et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309 (2001).
Cheng, Y., Dai, X. & Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799 (2006).
Box, M. S. et al. ELF3 controls thermoresponsive growth in Arabidopsis. Curr. Biol. 25, 194–199 (2015).
Jung, J. H. et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585, 256–260 (2020).
Yu, L. P., Miller, A. K. & Clark, S. E. POLTERGEIST encodes a protein phosphatase 2C that regulates CLAVATA pathways controlling stem cell identity at Arabidopsis shoot and flower meristems. Curr. Biol. 13, 179–188 (2003).
Song, S. K. & Clark, S. E. POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev. Biol. 285, 272–284 (2005).
Yu, L. P., Simon, E. J., Trotochaud, A. E. & Clark, S. E. POLTERGEIST functions to regulate meristem development downstream of the CLAVATA loci. Development 127, 1661–1670 (2000).
DeFalco, T. A. et al. A conserved module regulates receptor kinase signalling in immunity and development. Nat. Plants 8, 356–365 (2022).
Elliott, R. C. et al. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8, 155–168 (1996).
Clark, S. E., Running, M. P. & Meyerowitz, E. M. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067 (1995).
Shi, B. et al. Feedback from lateral organs controls shoot apical meristem growth by modulating auxin transport. Dev. Cell 44, 204–216.e6 (2018).
Ren, S. C. et al. CLE25 peptide regulates phloem initiation in Arabidopsis through a CLERK-CLV2 receptor complex. J. Integr. Plant Biol. 61, 1043–1061 (2019).
Durbak, A. R. & Tax, F. E. CLAVATA signaling pathway receptors of Arabidopsis regulate cell proliferation in fruit organ formation as well as in meristems. Genetics 189, 177–194 (2011).
Merelo, P., Gonzalez-Cuadra, I. & Ferrandiz, C. A cellular analysis of meristem activity at the end of flowering points to cytokinin as a major regulator of proliferative arrest in Arabidopsis. Curr. Biol. 32, 749–762.e3 (2022).
Vernoux, T. et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7, 508 (2011).
Wang, W. et al. Receptor-like cytoplasmic kinases PBL34/35/36 are required for CLE peptide-mediated signaling to maintain shoot apical meristem and root apical meristem homeostasis in Arabidopsis. Plant Cell 34, 1289–1307 (2022).
Wang, G. et al. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 147, 503–517 (2008).
Clark, S. E., Running, M. P. & Meyerowitz, E. M. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418 (1993).
Galvan-Ampudia, C. S. et al. Temporal integration of auxin information for the regulation of patterning. eLife https://doi.org/10.7554/eLife.55832 (2020).
Ma, Y. et al. WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat. Commun. 10, 5093 (2019).
Nemec-Venza, Z. et al. CLAVATA modulates auxin homeostasis and transport to regulate stem cell identity and plant shape in a moss. New Phytol. https://doi.org/10.1111/nph.17969 (2022).
Wu, C. C., Li, F. W. & Kramer, E. M. Large-scale phylogenomic analysis suggests three ancient superclades of the WUSCHEL-RELATED HOMEOBOX transcription factor family in plants. PLoS ONE 14, e0223521 (2019).
Lambolez, A. et al. Warm temperature promotes shoot regeneration in Arabidopsis thaliana. Plant Cell Physiol. https://doi.org/10.1093/pcp/pcac017 (2022).
Prigge, M. J. et al. Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. eLife https://doi.org/10.7554/eLife.54740 (2020).
Kinoshita, A. et al. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137, 3911–3920 (2010).
Dievart, A. et al. CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15, 1198–1211 (2003).
Hicks, K. A. et al. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274, 790–792 (1996).
Nimchuk, Z. L. & Perdue, T. D. Live imaging of shoot meristems on an inverted confocal microscope using an objective lens inverter attachment. Front. Plant Sci. 8, 773 (2017).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Barrell, P. J. & Conner, A. J. Minimal T-DNA vectors suitable for agricultural deployment of transgenic plants. Biotechniques 41, 708–710 (2006).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016).
Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013).
Risso, D., Schwartz, K., Sherlock, G. & Dudoit, S. GC-content normalization for RNA-seq data. BMC Bioinformatics 12, 480 (2011).
Risso, D., Ngai, J., Speed, T. P. & Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Thomas, P. D. et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003).
Peterson, B. A. et al. Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PLoS ONE 11, e0162169 (2016).
Acknowledgements
We thank T. D. Perdue, director of the University of North Carolina-Chapel Hill Genome Sciences Microscopy Core, for assistance with confocal imaging and A. Willoughby for assistance with SEM and photography; J. Winshell and J. Garzoni for lab and plant growth facility support; UNC’s High-Throughput Sequencing Facility for sequencing services; and members of the Nimchuk Lab for critical feedback on this project. This research was supported by a National Institute of General Medical Sciences—Maximizing Investigators’ Research Award from the NIH (R35GM119614, Z.L.N.), a National Science Foundation (NSF) Plant Genome Research Program (PGRP) grant (IOS-1546837, Z.L.N.), a National Science Foundation Postdoctoral Research Fellowship in Biology through the PGRP (NSF 1906389, D.S.J.) and a National Science Foundation Graduate Research Fellowship (DGE-2040435, E.S.S.). Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Author information
Authors and Affiliations
Contributions
A.J., E.S.S., D.S.J. and Z.L.N. conceptualized the project. A.J., E.S.S., D.S.J., C.L.S. and Z.L.N. developed the methodology. A.J., E.S.S., D.S.J. and C.L.S. performed validation. A.J., E.S.S., D.S.J. and C.L.S. conducted formal analysis. A.J., E.S.S., D.S.J. and C.L.S. conducted investigations. A.J., E.S.S. and D.S.J. curated the data. A.J., E.S.S., D.S.J. and Z.L.N. wrote the original draft. A.J., E.S.S., D.S.J. and Z.L.N. reviewed and edited the manuscript. A.J., E.S.S. and D.S.J. performed visualization. Z.L.N. acquired funding, supervised and administered the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Yvonne Stahl and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
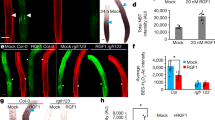
Extended Data Fig. 1 Temperature and CLV3p receptors intersect to buffer auxin mediated floral primordia formation.
a-e, CLE receptors and temperature intersect to buffer floral primordia formation. a, Quantification of flower primordia termination at 16 °C (n = 9–21), 23 °C (n = 10–24) and 30 °C (n = 12–15) for Col-0, pol, crn, crn pol, clv1-101, clv1-101 crn, clv1-101 pol, clv1-101 crn pol, cik124, cik124 crn, cik124 pol, cik124 crn pol. b, Inflorescence images at 16 °C and 30 °C of clv1-101, clv1-101 crn pol, cik124, and cik124 crn pol. All cik alleles in this figure are cik1-3, cik2-3, and cik4-3 CRISPR generated alleles. Same magnification within temperatures. c, Quantification of flower primordia termination at 16 °C (n = 8–9), 23 °C (n = 8–13), and 30 °C (n = 12) for Col-0, pol, crn, crn pol, clv1-8 crn, clv1-8 pol, clv1-8 crn pol. d, Inflorescence images at 16 °C, 23 °C and 30 °C of clv1-8 crn and clv1-8 crn pol. Same magnification within temperatures. e, Maximum intensity projection of DR5::GFP (green fire blue LUT) and PI (grey) and DR5::GFP alone in the IMs pol (n = 7), clv1-101 pol (n = 6), and clv1-101 crn (n = 8). f, Maximum intensity projection of Col-0 CLV1pro::2xYPET-N7 (green) inflorescence meristem. Expression patterns of CLV1pro::2xYPET-N7 in Col-0 (n = 4) and crn pol (n = 5) showing no expression changes. g, Expression patterns of POLpro::YPET-H2AX (green, n = 6) in Col-0 and WUSpro::POL-YPET (green, n = 3) and DR5::GFP (purple) in crn pol showing L5 and Z axis (PI, red). h, Diagram of flower/fruit production on primary inflorescence in wild-type and terminating and recovering crn plants. n = X biologically independent samples and statistical groupings based on significant differences using Kruskal-Wallis and Dunn’s multiple comparison test correction (a, c) where significance is defined as p-value < 0.05. Scale bars 10 mm (b, d) and 20 µm (e, f, g).
Extended Data Fig. 2 CIK co-receptors are required for temperature-dependent floral primordia outgrowth.
a, b, CIK receptors act redundantly in temperature sensitive floral primordia formation. a, Quantification of flower primordia termination at 16 °C (n = 7–14), 23 °C (n = 8–9) and 30 °C (n = 7–9) for Col-0, crn, cik1, cik2, cik3, cik4, cik12, cik13, cik14, cik234, cik124, cik134, cik123 and cik1234. b, Inflorescence images at 16 °C of cik1, cik12, cik13, cik14, cik124, cik134, cik123, cik1234 with arrows pointing to termination. Same magnification within single and double mutants as well as within triple and quadruple mutants. All cik alleles in this figure are cik1-1, cik2-1, cik3-1, and cik4-1 T-DNA lines. c. CIK genes are expressed in the IM. Expression patterns of YPET-H2AX (green fire blue LUT) reporter lines in the IM with XY view of L5 and axial view of same stack indicated with dotted line of CIK1pro (n = 10), CIK2pro (n = 15), CIK3pro (n = 8) and CIK4pro (n = 14) in Col-0 showing representative images (PI, magenta). n = X biologically independent samples and statistical groupings based on significant differences using Kruskal-Wallis and Dunn’s multiple comparison test correction (a) where significance is defined as p-value < 0.05. Scale bars 10 mm (b) and 20 µm (c).
Extended Data Fig. 3 CLV3p dependent stem fasciation and floral primordia outgrowth are temperature dependent.
a, Primordia outgrowth defects in clv3 plants are buffered by temperature. Quantification of termination at 16 °C (n = 8–21), 23 °C (n = 9–22), and 30 °C (n = 12–16) for Col-0, pol, crn, crn pol, clv3-20, clv3-20 crn, clv3-20 pol, clv3-20 crn pol. b, Stem fasciation in clv3 plants is buffered by temperature. Quantification of maximum stem width at 16 °C (n = 9), 23 °C (n = 9), and 30 °C (n = 15). c, Inflorescences at 16 °C and 30 °C of clv3-20, crn, clv3-20 pol and clv3-20 crn pol. Varying magnification is used at 16 °C to show clv3 fasciation and flower termination (arrows). All same magnification at 30 °C. n = X biologically independent samples and statistical groupings based on significant differences using a one-way ANOVA (a, 16 °C and 23 °C) and Kruskal-Wallis and Dunn’s multiple comparison test correction (a 30 °C and b) where significance is defined as p-value < 0.05. In box plots, box indicates interquartile zone (25–75th percentile) with median line at center, whiskers indicate minimum and maximum values with ‘+’ indicating mean (b). Scale bars 10 mm (c).
Extended Data Fig. 4 CLV-dependent FM stem cell regulation is sensitized to heat and auxin levels.
a, CLV-dependent FM stem cell regulation is repressed in warmer growth environments. Carpels per flower at 16 °C (blue) and 30 °C (red) with sample numbers as follow (16 °C, 30 °C) for Col-0 (n = 18, n = 8), pol (n = 25, n = 9), crn (n = 9, n = 14), clv1 (n = 9, n = 13), crn pol (n = 14, n = 11), clv1 pol (n = 11, n = 16), clv1 crn (n = 10, n = 15), clv1 crn pol (n = 10, n = 18), cik124 (n = 12, n = 9), cik124crn (n = 8, n = 16), cik124 pol (n = 13, n = 16), cik124 crn pol (n = 14, n = 17). Middle chart shows Col-0 (n = 18, n = 9), pol (n = 25, n = 9), crn (n = 9, n = 15), crn pol (n = 14, n = 14), clv3-20 (n = 8, n = 16), clv3-20 crn (n = 8, n = 13), clv3-20 pol (n = 10, n = 15), and clv3-20 crn pol (n = 12, n = 15). Last chart shows Col-0 (n = 9, n = 12), pol (n = 12, n = 12), crn (n = 9, n = 12), clv1-8 (n = 12, n = 12), crn pol (n = 9, n = 12), clv1-8 pol (n = 12, n = 12), clv1-8 crn (n = 9, n = 12), and clv1-8 crn pol (n = 12, n = 12). b, elf3 enhances carpel numbers in clv1-101 mutants. Carpels per flower at 16 °C (blue) and 30 °C (red) with sample numbers as follows (16 °C, 30 °C) for Col-0 (n = 10, n = 16), crn (n = 14, n = 12), elf3 (n = 14, n = 9), crn elf3 (n = 13, n = 10), clv1 (n = 9, n = 11), clv1-101 elf3 (n = 13, n = 12), clv1 crn (n = 11, n = 14) and clv1-101 crn elf3 (n = 14, n = 12) with all single mutant control alleles. c, CLV-mediated FM stem cell regulation is sensitized to auxin levels. Carpels per flower at 23 °C for Col-0 (n = 9), yuc4 (n = 7), clv1-101 (n = 8), clv1-101 yuc4 (n = 8), clv3-9 (n = 11), clv3-9 yuc4 (n = 12). In box plots, box indicates interquartile zone (25-75th percentile) with median line at center, whiskers indicate minimum and maximum values with ‘+’ indicating mean (a-c). Statistical comparisons based on two-tailed Mann-Whitney test (a) where * indicate p-value < 0.0001 and ns indicates not significant. n = X biologically independent samples and statistical groupings based on significant differences using a Kruskal-Wallis and Dunn’s multiple comparison test correction (b, c). Significance is defined as p-value < 0.05 (a-c).
Extended Data Fig. 5 CLV3 acts redundantly with CLE25 and auxin in IM function.
a-c, cle25 and yuc1/4 mutations enhance clv3 IM phenotypes. a, SEM micrographs of IMs of cle26-10 clv3-20, cle27-10 clv3-20, cle26-10 cle27-10 clv3-20, clv3-20, cle25-10 clv3-20, cle25-10 cle26-10 clv3-20, cle25-10 cle27-10 clv3-20, and cle25-10 cle26-10 cle27-10 clv3-20. clv3-9 yuc14 and clv3-9 inset at same magnification for size comparison. clv3-9 yuc1/4 side view. Scale bars 200 µm and representative images shown from 4 independent experiments. b, Flower primordia termination visualized with SEM side view of IMs in Col-0, crn, clv3-9, clv3-20, cle26-10 cle27-10 clv3-20, cle25-10 clv3-20, cle25-10 cle26-10 cle27-10 clv3-20. Scale bars 1 mm. Representative images from 2 independent experiments shown. c, Inflorescence of the cle25-11 allele with clv3-20 mutation showing characteristic disk-like IM phenotype. d, Quantification of disk meristem expansion using ratio of maximum width and maximum length of clv3-20 (n = 12), clv3-20 crn (n = 9), clv3-9 yuc1/4 (n = 9), cle25-10 clv3-20 (n = 14), cle26-10 cle27-10 clv3-20 (n = 8) and cle25-10 cle26-10 cle27-10 clv3-20 (n = 10). e, CRISPR guide and PAM sites for CLE genes indicating SNPs and indels in red or with dash. WT, wild-type; m, mutant. f, Gene diagrams of CLE ORFs with red line indicating CRISPR target positioned before the CLE domain indicated by the blue rectangle. g, carpels per flower of Col-0, cle25-10, clv3-20 and cle25-10 clv3-20 (n = 12). h-i, cle25 alone does not enhance crn. h, Inflorescence images (scale bars 10 mm) and i, quantification of termination of Col-0, cle25-10, crn and cle25-10 crn (n = 9). CLE25 is expressed in root phloem cells independent of clv3. j, Confocal images of CLE25pro::YPET-H2AX (green) of Col-0 and clv3-20 in roots. f, CLE25 is ectopically expressed in clv3-20 floral primordia. Col-0 (n = 4) and clv3-20 (n = 4) with same single transgene CLE25pro::YPET-H2AX (green) insertion line (#8) in IM and FMs (PI, magenta). Scale bar 20 µm (j, k). In box plots, box indicates interquartile zone (25–75th percentile) with median line at center, whiskers indicate minimum and maximum values with ‘+’ indicating mean (d, g). n = X biologically independent samples and statistical groupings based on significant differences using a Kruskal-Wallis and Dunn’s multiple comparison test correction (d, g, i) where significance is defined as p-value < 0.05.
Extended Data Fig. 6 CLV1 is required for IM maintenance.
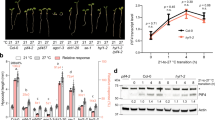
a-e, CLV1 is required for primary IM maintenance. a, Side view of primary inflorescence with arrows pointing to primary inflorescence (white) with cauline leaf (green) subtending the secondary inflorescence (yellow) in both Col-0 and clv1-101 revealing primary inflorescence termination (PIT) in clv1-101. b, Categories: buried, hook, weak, WT-like, and fasciated. Range of flowers that emerge in each category before PIT. c, Population analysis of clv1-101 at 16 °C (n = 87). d, UBQ10::DII-Venus in L1 cells of the IM in clv1-101/+ and clv1-101 homozygous mutant undergoing PIT. The same transgenic insertion line was used in d. 4/4 WT UBQ10::DII-Venus and 4/5 clv1-101 UBQ10::DII-Venus (green) independent transgenic lines showed representative expression. clv1-20 allele inflorescence undergoing PIT at 23 °C but not at 30 °C (PI, magenta) (e) and quantification of PIT in Col-0, clv1-101, clv1-20 and clv1-8 plants (f, n = 15). g, Summary of interactions between CLEp signaling and heat on auxin dependent stem cell regulation, flower primordia outgrowth, and SAM maintenance. n = X biologically independent samples. Scale bars 10 mm (b, d, e) and 20 µm (d).
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and Tables 1 and 2.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
John, A., Smith, E.S., Jones, D.S. et al. A network of CLAVATA receptors buffers auxin-dependent meristem maintenance. Nat. Plants 9, 1306–1317 (2023). https://doi.org/10.1038/s41477-023-01485-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-023-01485-y