Abstract
In situ mRNA hybridization is one of the most powerful techniques for analyzing patterns of gene expression. However, its usefulness is limited in complex plant tissues by the need to fix, embed and section samples before localizing the desired mRNA. Here we present a robust whole-mount in situ hybridization method that allows easy access to patterns of gene expression in intact, complex tissues, such as the inflorescence apex of Arabidopsis thaliana. The tissue is first fixed and then permeabilized by treatment with a cocktail of cell wall–digesting enzymes that has been optimized to preserve the integrity of tissue structures, while also permitting the detection of expression patterns in deep tissues. In addition to colorimetric staining, fluorimetric staining that can be imaged by confocal microscopy can also be used with this protocol, thus providing full 3D resolution. The entire procedure can take <3 d from tissue preparation to image acquisition.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cox, K.H., DeLeon, D.V., Angerer, L.M. & Angerer, R.C. Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev. Biol. 101, 485–502 (1984).
Jackson, D. in Molecular Plant Pathology: A Practical Approach (ed. Bowles, D.J., Gurr, S.J. & McPherson, M.) (Oxford University Press, 1991).
Tautz, D. & Pfeifle, C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98, 81–85 (1989).
Hejátko, J. et al. In situ hybridization technique for mRNA detection in whole-mount Arabidopsis samples. Nat. Protoc. 1, 1939–1946 (2006).
Leduc, N., Matthys-Rochon, E. & Dumas, C. Deleterious effect of minimal enzymatic treatments on the development of isolated maize embryo sacs in culture. Sex. Plant Reprod. 8, 313–317 (1995).
Matthys-Rochon, E., Mòl, R., Heizmann, P. & Dumas, C. Isolation and microinjection of active sperm nuclei into egg cells and central cells of isolated maize embryo sacs. Zygote 2, 29–35 (1994).
Besnard, F. et al. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 505, 417–421 (2014).
Milani, P. et al. Matching patterns of gene expression to mechanical stiffness at cell resolution through quantitative tandem epifluorescence and nanoindentation. Plant Physiol. 165, 1399–1408 (2014).
Besnard, F., Rozier, F. & Vernoux, T. The AHP6 cytokinin signaling inhibitor mediates an auxin-cytokinin crosstalk that regulates the timing of organ initiation at the shoot apical meristem. Plant Signal. Behav. 9, pii: e28788 (2014).
Clark, S.E., Running, M.P. & Meyerowitz, E.M. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067 (1995).
Mayer, K.F. et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815 (1998).
Sauer, M., Paciorek, T., Benkova, E. & Friml, J. Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat. Protoc. 1, 98–103 (2006).
Lauter, G., Söll, I. & Hauptmann, G. Multicolor fluorescent in situ hybridization to define abutting and overlapping gene expression in the embryonic zebrafish brain. Neural Dev. 6, 10 (2011).
Fernandez, R. et al. Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nat. Methods 7, 547–553 (2010).
Fletcher, J.C., Brand, U., Running, M.P., Simon, R. & Meyerowitz, E.M. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914 (1999).
Drews, G.N., Bowman, J.L. & Meyerowitz, E.M. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65, 991–1002 (1991).
Acknowledgements
We thank A. Lacroix and P. Villand for help with plant growth, C. Lionnet (PLATIM imaging platform) for help with imaging and J.H. Doonan for his insights. P.D. was supported by an INRA Chaire d'excellence grant, and T.V. was supported by the ANR-12-BSV6-0005 (AuxiFlo) grant.
Author information
Authors and Affiliations
Contributions
T.V. and P.D. designed and supervised experiments; F.R. performed the experiments with the help of V.M. and P.D.; F.R., V.M., T.V. and P.D. analyzed the data; and F.R., V.M., T.V. and P.D. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
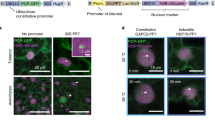
Supplementary Figure 1 Expected results for variations in the tissue permeabilization and probe hydrolysis steps.
(a–b) Examples of whole-mount in situ experiments to localize WUSCHEL mRNA in tissue that has been subjected to enzymatic permeabilization of either 0 min (a) or 6 min (b). Compare the absent or reduced signal in the thicker parts of the tissue, such as the center of the SAM (red arrowheads) or in older flowers, either at stage 3 (blue arrows) or stage 2 (red arrows). (c) Lane 1 shows the products of a transcription reaction (for 1 μl of the 20 μl reaction volume) from a PCR-amplified full-length (291 bp) CLAVATA3 template. Note the presence of an additional larger band (solid triangle), which is visible in most transcription reactions. Lane 2 shows the results of a successful hydrolysis reaction (for 5 μl of the 80-100 μl reaction volume), where a smear centered at roughly 0.1 kb is typically observed (open triangle). In the case of an unsuccessful hydrolysis, lane 2 would look identical to lane 1.
Supplementary information
Supplementary Figure 1
Expected results for variations in the tissue permeabilization and probe hydrolysis steps. (PDF 502 kb)
Rights and permissions
About this article
Cite this article
Rozier, F., Mirabet, V., Vernoux, T. et al. Analysis of 3D gene expression patterns in plants using whole-mount RNA in situ hybridization. Nat Protoc 9, 2464–2475 (2014). https://doi.org/10.1038/nprot.2014.162
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.162
This article is cited by
-
Decoding the leaf apical meristem of Guarea glabra Vahl (Meliaceae): insight into the evolution of indeterminate pinnate leaves
Scientific Reports (2024)
-
A rapid and sensitive, multiplex, whole mount RNA fluorescence in situ hybridization and immunohistochemistry protocol
Plant Methods (2023)
-
A tool for multiplexed spatial gene expression analysis in plant tissues
Nature Plants (2023)
-
Visualization of spatial gene expression in plants by modified RNAscope fluorescent in situ hybridization
Plant Methods (2020)
-
Genetic control of meristem arrest and life span in Arabidopsis by a FRUITFULL-APETALA2 pathway
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.