Abstract
Recent identification of somatic MED12 mutations in most uterine leiomyomas brings a new venue for the study of the tumorigenesis of leiomyomas. We are particularly interested in the correlation of MED12 and HMGA2 gene products in leiomyomas and leiomyosarcomas with and without MED12 mutations. To address these issues, in this study we examined MED12 mutations in a large cohort of usual type leiomyomas (178 cases) and uterine leiomyosarcomas (32 cases). We found that 74.7% (133/178) of leiomyomas had MED12 mutations, which was consistent with several independent studies. In contrast, only 9.7% (3/32) of leiomyosarcomas harbored MED12 mutations. Expression analysis by western blot and immunohistochemistry revealed that those leiomyomas with complex MED12 mutations had significantly lower protein products than the matched myometrium. Interestingly, most leiomyosarcomas without MED12 mutations also had very low levels of MED12 expression in comparison to the matched myometrium. These findings suggest a potential functional role of MED12 in both benign and malignant uterine smooth muscle tumors. When we further examined HMGA2 expression in all leiomyomas and leiomyosarcomas, we found that HMGA2 overexpression was exclusively present in those leiomyomas with no MED12 mutation, accounting for 10.1% (18/178) of total leiomyomas and 40% (18/45) of non-MED12 mutant leiomyomas. Twenty-five percent (8/32) of leiomyosarcomas had HMGA2 overexpression, and no MED12 mutations were found in HMGA2 positive leiomyosarcoma. These findings strongly suggest that MED12 mutations and HMGA2 overexpression are independent genetic events that occur in leiomyomas, and they may act differently in the tumorigenesis of uterine leiomyomas.
Similar content being viewed by others
Main
The causes of uterine leiomyomas have been investigated for many years. Although some genetic mutations have been identified in a small fraction of leiomyomas,1, 2, 3, 4, 5 some of them have been confirmed to be primary genetic events specifically of germline fumarate hydratase (FH) mutations.6 It was not until recently that Mäkinen et al,7 identified a high rate of somatic gene mutations in the mediator complex subunit 12(MED12) gene, based on the whole exome sequencing analysis. This group further examined a total of 207 leiomyomas and nearly 70% of them had MED12 mutations.7 Immediately following this discovery, two additional large cohort mutation analyses in human uterine leiomyomas were conducted and had similar findings.8, 9 Current studies reveal that MED12 mutations/variants in leiomyomas: (1) occur with a high frequency (up to 70% of tumors harbor MED12 mutations); (2) are site-specific (71% of mutations are in codon 44, exon 2 of MED12); and (3) are heterozygous with no mutation detected in the normal myometrium.
In a study of 1862 tumor samples of mesenchymal and epithelial neoplasms, MED12 mutations/variants are specific to uterine leiomyomas and are rarely found in epithelial neoplasms.10 As uterine smooth muscle tumors consist of a group of heterogeneous tumors, including different histological variants, the role of MED12 mutations in different types of uterine smooth muscle tumors is interesting but remains largely unknown. Examination of MED12 mutations in different uterine smooth muscle tumors9, 11 may help to disclose the relationship between usual type leiomyomas and other histologic variants or their malignant counterpart. Leiomyosarcomas are traditionally considered to arise de novo; however, recent studies suggest that some leiomyosarcomas may arise from existing leiomyomas.12, 13 The latter hypothesis is attractive, but still lacks direct molecular support. Findings of MED12 mutations in 2–20% of uterine leiomyosarcoma11, 14, 15, 16, 17 suggest that at least some leiomyosarcoma share the same sequence of MED12 mutations.
About one third of uterine leiomyomas have no MED12 mutations, and we want to study the role of HMGA2 in leiomyomas without MED12 mutations. We also want to know whether MED12 mutations can be used as a biomarker for differential diagnosis. To address these issues, we first examined the spectrum of MED12 mutations in a large cohort of benign and malignant uterine smooth muscle tumors. Then, we compared the correlation of gene mutations with the protein expression and further analyzed the patterns between MED12 mutations and HMGA2 expression in all of the tumors. Our results indicate that MED12 mutations are common in benign but much less common in malignant uterine smooth muscle tumors. In particular, we have demonstrated that genetic mutations of MED12 and HMGA2 do not overlap in any given leiomyoma; therefore, our findings strongly suggest the presence of two independent genetic pathways in the tumorigenesis of uterine leiomyomas.
Materials and methods
Case Selections
The use of human tissue specimens was approved by the Institutional Review Board for Human Research at Northwestern University, Chicago, IL. Fresh frozen and/or formalin-fixed and paraffin-embedded tumor, and myometrial tissues were obtained from female patients (range: 22–56 years) undergoing hysterectomies or myomectomies at the Prentice Women’s Hospital in Chicago IL. Before surgery, written informed consent and demographic/endocrine information (eg, race, age, day of menstrual cycle, parity, whether oral contraception is being taken) was obtained. A total of 178 cases with histologic diagnosis of usual type leiomyoma and 32 cases with uterine leiomyosarcoma were selected for the study. The diagnoses of leiomyosarcomas were based on Bell criteria18 and clinical presentation. Patient’s clinical biodemography and pathological features were summarized in Table 1.
Immunofluorescence and Confocal Image
Immunofluorescence was performed as previously described.19 Briefly, human myometrium and leiomyoma tissues were fixed in a modified Davidson’s solution (Electron Microscopy Science, Hatfield, PA), embedded in paraffin, and sectioned at 5 μm. Anti-MED12 (1:200, Proteintech Europe, Manchester, UK) and αSMA (1:50, Abcam, Cambridge, MA) were used as primary antibodies. Alexa Fluro 488 goat anti-mouse and Alexa Fluro 594 goat anti-rabbit (Molecular Probes/Invitrogen) were used as secondary antibodies. For the counterstaining, sections were incubated with 150 ng/ml DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) (Sigma-Aldrich) for 5 min at room temperature. Confocal images were obtained with an Olympus Fluoview FV10i Confocal Microscope system (Olympus America Inc, Center Valley, PA).
Tissue Microarray and Immunohistochemistry
Tissue cores were collected from tumors and myometrial controls of each case for tissue microarray were represented in duplicate. Antibodies used for this study included HMGA2 (1:500, BioChem Inc, CA, USA ) and Anti-MED12 (1:200, Proteintech Europe, Manchester, UK). Tissue microarrays were sectioned, 4 μm in thickness. After deparaffinization and antigen retrieval, all immunohistochemical staining was performed on a Ventana Nexus automated system (Tucson, AZ, USA). In brief, the endogenous peroxidase activity was blocked with 3% hydrogen peroxide. Primary antibodies were detected using standard biotinylated anti-mouse or anti-rabbit secondary antibodies. Semi-quantative immunointensity was scored as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong) and percentage was scored as 0(0%), 1(≤10%), 2 (11–50%), 3(51–80%) ,and 4(>80%). Immunoreactivity for MED12 was scored for intensity only as it is diffusely expressed in all positive cases. The majority of HMGA2 positive cases showed strong and diffuse immunoreactivity for HMGA2, and only those cases with weak or faint immunoreactivity (intensity score 1) for HMGA2 showed a patchy staining pattern. Therefore, a combined intensity and percentage of immunoreactivity for HMGA2 were used. Moderate to high immunoreactivity (≥2) for HMGA2 in >50% of nuclei was read as HMGA2 positive.
Western Blotting
Freshly frozen myometrium and leiomyoma tissues were homogenized at 4 °C in a protein lysis buffer. Total cellular proteins from cultured cell samples were prepared with a modified radioimmunoprecipitation assay buffer, as described previously.20 Protein concentrations were determined with the BCA protein assay kit (Pierce Protein Research Products, Rockford, IL, USA). The cell lysates (40 μg of total protein per sample) were separated on 8% sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gels. After electrophoresis, the proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore Corp, Bedford, MA, USA). The blotted membranes were blocked for 1 h in a Tris-buffered saline with 5% non-fat milk at room temperature and subsequently incubated with primary antibodies against the proteins of interest. The rabbit anti-human MED12 was obtained from Bethyl Laboratory (Montgomery, TX, USA ). After incubation with the primary antibody, the PVDF membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibodies (goat anti-rabbit, Amersham Biosciences, Piscataway, NJ, USA). Immune complexes were detected on the membranes with a chemiluminescent reagent (GE Healthcare Biosciences, Piscataway, NJ, USA). An anti-β-actin monoclonal antibody (Abcam, Cambridge, MA, USA) and an anti-α-smooth muscle actin (Abcam) or anti-GAPDH (Cell signaling technology, Danvers, MA, USA) were used as a loading control. Net intensities for the protein bands of interest were determined using Adobe Photoshop. Band intensities were then normalized to β-actin intensities and compared for differences between experimental and control conditions.
DNA Extraction from Fresh Frozen and Formalin-Fixed and Paraffin-Embedded Tissue
We used the Qiagen DNeasy Blood and Tissue kit and the protocols for fresh frozen and formalin-fixed tissues. For formalin-fixed and paraffin-embedded tissue, six to eight 10 μm tissue sections were collected into 1.5 ml Ependoff tubes. Paraffin was dissolved in xylene and removed. Tissue samples were then lysed under denaturing conditions with a proteinase K digestion at 56 °C for 1 h, followed by incubation at 90 °C to reverse formalin cross-linking. DNA was purified by column purification with a filter membrane and stored in −20 °C before use.
PCR and Sequencing
For PCR amplification, 50 ng of genomic DNA was used. Primers used for the amplification of MED12 exon 2 and flanking the exon–intron junction sequence were 5′- GCC CTT TCA CCT TGT TCC TT-3′ (forward) and 5′-TGT CCC TAT AAG TCT TCC CAA CC-3′ (reverse). PCR products were purified by ExoSAP-IT reagent (Affymetrix, Inc) following the manufacturers instruction. DNA sequencing of the purified DNA products was performed by GENEWIZ, Inc (South Plainfield, NJ, USA) and Genomic Core Facility at the Northwestern University using Applied Biosystems BigDye version 3.1. The reactions were then run on Applied Biosystem’s 3730xl DNA Analyzer. Mutations/variations were analyzed by the DNASTAR Lasergene 9 software for analysis.
Statistical Analysis
Gene expression levels by immunohistochemistry were summarized by medians and ranges for the entire patient population, and separated by tumors and matched myometrium, and histology and clinical groups. Differences between tumors and matched myometrium, and among histology and clinical groups were assessed using Wilcoxon rank-sum tests. Associations among the gene expression levels were assessed via the Spearman correlation. P values less than 0.05 (P<0.05) were considered statistically significant.
Results
MED12 Mutations in Usual Type Leiomyomas and Leiomyosarcomas
We collected 178 uterine leiomyomas, and all usual type reviewed and confirmed by histology examination. Tumor size ranged from 3 to 21 cm. Patients’ age ranged from 22 to 56 years with a median age of 42 years old (Table 1). Among them, 64 were black women, 52 white women, 13 Hispanic, and four Asian women (Table 2).
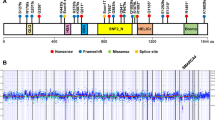
Of all 178 leiomyomas examined, 74.7%(133/178) of uterine leiomyomas had MED12 mutations. Among them, 93.9%(125/133) of mutations were confined to exon 2 and the remaining 6.1% in intron 1. For those leiomyomas with MED12 mutations, point mutations were found in 87.9%(117/133) of cases and complex MED12 mutations in 12.1%(16/133). Overall, point mutations spread to 12-single nucleotide sites, and the most common point mutations were found in two nucleotide sites of c130 and c131, accounting for 72.9%(97/133) of MED12 mutations (Figure 1, Table 1). Leiomyomas with MED12 mutations seemed to be evenly distributed among different races, and no racial difference was noted (Table 1). This is consistent with another recent study.9 No significant associations of MED12 mutations with tumor size and patients age were seen.
MED12 mutation analysis in uterine leiomyomas. (a) The photomicrograph illustrates the histology of usual type leiomyoma. (b) Sequencing chromatographs of 12 types of MED12 mutations identified in usual type leiomyomas. Red arrows indicate the mutation sites. (c) Genomic analysis of the types and distribution of MED12 mutations in exon2 and intron1. The point mutations were listed in the above sequences and each dot represents a case with mutation. The complex mutations were listed below the sequence with thin bars illustrating the actual length of deletions.
A total of 32 uterine leiomyosarcomas were included in this study. Patients age ranged from 47 to 86 years old and the tumor size of 2.3–18.6 cm (Table 1). Fifty-nine percent of them were stage I and the rest of the cases were II–IV. Only three cases presented with MED12 mutations accounting for 9.4% of the tumor population (3/32). Of three cases with MED12 mutations, one had a deletion of 39 nucleotides, corresponding to 13 amino acids shorter, one had a frame-shift likely resulting in no protein product, and one had a point mutation of G–A change at a position of 131 (Figure 2). Findings suggest that MED12 mutations in uterine leiomyosarcomas are very low.
Expression Analysis of Wild Type and Mutant MED12 in Leiomyomas and Leiomyosarcomas
The correlation of MED12 mutations and its gene product in leiomyoma remains unclear. Studying MED12 gene products will not only help us to understand the function of this gene but will also provide a potential biomarker for clinical application, as well as for differential diagnosis. To this end, we examined MED12 gene products by western blot in the selected leiomyomas with wild type, simple, and complex MED12 mutations. Simple mutations were defined as point mutations involving a single nucleotide replacement, whereas complex MED12 mutations referred to those changes of insertion, deletion, and shifting of the coding sequence (Figure 1). We found that leiomyomas with a complex MED12 mutation seemed to have very low or a complete loss of its gene product, and those tumors with simple MED12 mutations showed slightly reduced protein expression (Figure 3). These findings were further supported by the immunofluorescent stain of MED12. With the aid of internal counterstains of DAPI and smooth muscle actin-α, loss of MED12 expression in leiomyomas with complex MED12 mutations was evident in comparison to its matched myometrial tissue section (Figure 3).
Reduced MED12 protein levels in uterine leiomyomas with complex MED12 mutations. (a) Confocal microscopic images illustrate an example of immunofluorescent staining for MED12 (red), smooth muscle actin (green), and DAPI (blue) in leiomyoma with a complex MED12 mutation (with a partial MED12 Exon 2 deletion) and its matched myometrium. In myometrium, the expression pattern and distribution are punctuated with high intensity staining enriched in perinuclear localization. In contrast, the expression pattern for leiomyoma is diffused in the cytoplasm and has lesser staining intensity. Scale bars=50 μm. Higher power image of MED12 staining in leiomyoma and myometrium are shown in the insert (Scale bar=25 μm). (b) The expression of MED12 protein was detected by western blot analysis in leiomyomas with complex MED12 mutations (Pt81LM and Pt151LM for Exon 2 deletion on right) and leiomyomas with simple MED12 point mutations (Pt137LM and Pt173LM for c.131G>A on left). MED12 expression in the matched myometrium (MM) was shown next to tumors. Anti-α-smooth muscle actin, Anti-Actin, and anti-GAPDH were used as protein loading controls.
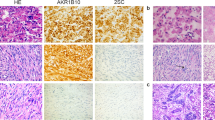
To explore the potential application of immunohistochemistry for MED12 expression in a formalin-fixed and paraffin-embedded tissue, we examined MED12 expression by immunohistochemistry (see Materials and methods) in all 178 leiomyomas and matched myometrium in a tissue microarray. As immunoreactivity for MED12 seemed to be diffusely distributed in the cytoplasm with some nuclear localization, we scored immunointensity by semiquantitative scales of negative, weak, moderate, and strong expression (Figure 4). Consistent with western blot results, we found that leiomyomas with complex MED12 mutations had significant lower immunoreactivity for MED12 (P<0.01) than in myometrium. Notably, no significant difference of MED12 expression between myometrium and leiomyomas of wild type and simple MED12 mutations was found.
Immunohistochemical analysis of MED12 expression in formalin-fixed and paraffin-embedded leiomyomas and leiomyosarcomas. (a) Photomicrographs illustrate examples of immunointensity of MED12 in tumor myometrium (MM), leiomyomas (ULM) and leiomyosarcoma (LMS). wt: wild type MED12; point: point/single nucleotide mutation of MED12; complex: complex mutations of MED12. (b) Statistical analysis of immunointensity for MED12 in tumor myometrium (MM), leiomyomas (ULM) and leiomyosarcoma (LMS). MED12 mutation types and number of cases were listed below the histobars. **P<0.01.
As reported, only three leiomyosarcomas had MED12 mutations. However, most leiomyosarcomas without the MED12 mutations showed significantly lower, or were absent of, immunoreactivity for MED12 (Figure 4).
HMGA2 Expression in Leiomyomas and Leiomyosarcoma with and without MED12 Mutations
Twenty-six percent of leiomyomas had no MED12 mutations. HMGA2 (high mobility group A2) overexpression induced by non-random chromosomal translocation t(12;14) was found in less than 10% of leiomyomas.21 To investigate whether HMGA2 overexpression correlates with MED12 mutations, we examined and compared the expression pattern of HMGA2 with MED12 mutations by immunohistochemistry in all 178 leiomyomas and 32 leiomyosarcomas. We found that a total of 18 leiomyomas and eight leiomyosarcomas were strongly immunoreactive for HMGA2, accounting for 10.1 and 25.0% of cases, respectively. Surprisingly, HMGA2 overexpression was exclusively found in those leiomyomas without a MED12 mutation (Figure 5). To confirm this finding, we repeated the HMGA2 stain in a different lab using a new vial of HMGA2 antibody of BioCheck, and the same results were found. Consistent with the finding in leiomyomas, leiomyosarcomas with HMGA2 overexpression did not harbor the MED12 mutation either. Our result was further supported by a study of 38 leiomyomas, in which the MED12 mutation and HMGA2 overexpression were identified in completely separate tumor populations.22
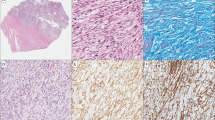
HMGA2 overexpression in uterine leiomyomas. (a) Photomicrographs illustrate examples of strong and diffuse immunoreactivity for HMGA2 in uterine leiomyomas. High grade serous ovarian carcinoma was used as the positive control (left upper). (b) and (c) Dot-plot analysis of distribution of MED12 mutations and HMGA2 overexpression in 178 cases of uterine leiomyomas. Seventy-four percent of leiomyomas have MED12 mutations and lack of HMGA2 overexpression; 10% of leiomyomas have wild type MED12 and HMGA2 overexpression; and remainder 16% of leiomyomas have wild type MED12 and lack of HMGA2 overexpression.
Discussion
Uterine leiomyoma is a major public health problem. Millions of reproductive aged women suffer from these tumors, and there are roughly 200 000 hysterectomies for symptomatic leiomyomas performed annually in the United States23 causing substantial medical costs.24, 25 Uterine leiomyomas are genetically heterogamous. Some tumors harbor non-random chromosomal translocations t(12;14) for HMGA2 overexpression,26, 27 and rare cases have germline gene mutations in FH28 and COL4A5/A6.29 Recent identification of MED12 mutations in most uterine leiomyomas provides a new and promising candidate gene for understanding the cause and tumorigenesis of leiomyomas. In particular, MED12 mutations seem to be mostly confined to uterine leiomyomas.13, 17 Although it is still too early to say that MED12 mutations are the direct causal factor for the development of most leiomyomas, these clinically significant findings will accelerate medical research to characterize the functional role of MED12 in the disease. Eventually, this will benefit the clinical application.
Based on several large cohorts and this study, the mutation rate of MED12 in uterine leiomyomas varies with a range from 50 to 70% of tumor population. MED12 mutations in uterine leiomyomas were mostly identified in exon 2 and rarely in the intron 1–exon 2 junction.7, 8 All mutations were heterozygous in genomic DNA, and all of the transcripts were derived solely from the mutant MED12 alleles.10, 11 Therefore, in this study, we examined MED12 gene mutations covering the exon 2 and intron 1–exon 2 junction. We found that 75% of tumors harbor MED12 mutation (Figure 1).
Notably, at least a quarter of leiomyomas have no detected MED12 mutations. HMGA2 overexpression is the second most common genetic alteration in leiomyomas, identified in 7.5–10% of leiomyomas due to t(12;14) translocation. In two independent studies, a total of 118 leiomyomas with cytogenetic information,9, 22 and no MED12 mutations in tumors with HMGA2 overexpression were found. To further investigate the relationship of MED12 mutations and HMGA2 overexpression, we examined HMGA2 expression in all 178 leiomyomas and 32 leiomyosarcomas by immunohistochemistry. As illustrated in Figure 5, HMGA2 overexpression was exclusively found in those tumors without MED12 mutations. The current study is the largest sample size for the analysis of MED12 mutations and HMGA2 expression. Combining these three studies into nearly 300 cases strongly suggests that there are two mutually exclusive independent pathways of MED12 and HMGA2, and they are likely playing different roles in the tumorigenesis of uterine leiomyoma.
Furthermore, although there is a low rate of MED12 mutations in uterine leiomyosarcomas, these two genetic mutations were independent. HMGA2 is an oncogene, and its oncogenic properties have been well characterized in many benign and malignant neoplasms. A recent study further defined that HMGA2-mediated tumorigenesis in leiomyomas is mainly through a regulation of the cell-cycle checkpoint in the G1/S phase.21 In the current study, we found that HMGA2 overexpression accounts for 45% of MED12 negative tumors. The findings suggest that some other genetic mutations, such as HMGA1, FH, COL4A5/A6, and as yet to be characterized genetic factors may be responsible for the remaining leiomyomas without HMGA2 and MED12 mutations.
As mentioned above, MED12 mutations are the major genetic changes in benign leiomyoma, whereas the mutations are much less common in the malignant counterpart—leiomyosarcoma. Furthermore, MED12 protein expression in leiomyomas with MED12 mutations has only been examined by the western blot analysis in one study,16 and no immunohistochemistry analysis for MED12 has been reported. To assess the value of immunohistochemistry for MED12 expression in formalin-fixed paraffin-embedded tissues, we compared and correlated the immunointensity of MED12 in tumors with/without MED12 mutations. Although western blot analysis can detect the reduced MED12 protein in both simple (point) and complex mutations of the MED12 gene (Figure 3), the immunohistochemical stain can only detect the reduced immunoreactivity for MED12 in tumors with the complex MED12 mutations (Figure 4). More studies are needed to further optimize the immunohistochemistry stain of this important gene marker for its potential clinical application.
MED12 mutations in leiomyosarcoma have drawn great attention recently and several studies have shown that MED12 mutations in leiomyosarcoma ranged from 3 to 20%.11, 14, 17 In this study, we found that the rate of MED12 mutations was nearly 10% in uterine leiomyosarcomas. When we combined all of the studies of leiomyosarcomas reported in the literature with our cases,10, 14, 15, 16, 17, 26 a total of 174 leiomyosarcomas, 14 tumors had MED12 mutations, accounting for an 8.1% mutation rate. Therefore, MED12 mutations in leiomyosarcoma are far lower than those in leiomyomas. Interestingly, two of three MED12 mutations in our series are either large deletion or frame-shift, which are rarely seen in leiomyomas (Figure 2). Nevertheless, 9.4% of leiomyosarcomas share genetic mutations of MED12 with leiomyomas, suggesting that a small fraction of leiomyosarcomas may progress from existing leiomyomas. This may warrant further study.
In summary, our current study of benign and malignant uterine smooth muscle tumors concludes that: (1) MED12 mutations are common in usual type leiomyomas; (2) leiomyomas with HMGA2 overexpression do not share or harbor MED12 mutations; (3) complex MED12 mutations lead to loss or reduced MED12 protein product; and (4) leiomyosarcomas have a very low rate of MED12 mutations, suggesting that most leiomyosarcomas have independent tumorigenic pathways from their benign counterpart.
References
Stewart EA, Morton CC . The genetics of uterine leiomyomata: what clinicians need to know. Obstet Gynecol 2006;107:917–921.
Gross KL, Neskey DM, Manchanda N et al. HMGA2 expression in uterine leiomyomata and myometrium: quantitative analysis and tissue culture studies. Genes Chromosomes Cancer 2003;38:68–79.
Hennig Y, Rogalla P, Wanschura S et al. HMGIC expressed in a uterine leiomyoma with a deletion of the long arm of chromosome 7 along with a 12q14-15 rearrangement but not in tumors showing del(7) as the sole cytogenetic abnormality. Cancer Genet Cytogenet 1997;96:129–133.
Rein MS, Powell WL, Walters FC et al. Cytogenetic abnormalities in uterine myomas are associated with myoma size. Mol Hum Reprod 1998;4:83–86.
Hennig Y, Deichert U, Bonk U et al. Chromosomal translocations affecting 12q14-15 but not deletions of the long arm of chromosome 7 associated with a growth advantage of uterine smooth muscle cells. Mol Hum Reprod 1999;5:1150–1154.
Tomlinson IP, Alam NA, Rowan AJ et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet 2002;30:406–410.
Makinen N, Mehine M, Tolvanen J et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 2011;334:252–255.
McGuire MM, Yatsenko A, Hoffner L et al. Whole exome sequencing in a random sample of North American women with leiomyomas identifies MED12 mutations in majority of uterine leiomyomas. PLoS One 2012;7:e33251.
Markowski DN, Bartnitzke S, Loning T et al. MED12 mutations in uterine fibroids-their relationship to cytogenetic subgroups. Int J Cancer 2012;131:1528–1536.
Je EM, Kim MR, Min KO et al. Mutational analysis of MED12 exon 2 in uterine leiomyoma and other common tumors. Int J Cancer 2012;131:1044–1047.
Perot G, Croce S, Ribeiro A et al. MED12 alterations in both human benign and malignant uterine soft tissue tumors. PLoS One 2012;7:e40015.
Mittal KR, Chen F, Wei JJ et al. Molecular and immunohistochemical evidence for the origin of uterine leiomyosarcomas from associated leiomyoma and symplastic leiomyoma-like areas. Mod Pathol 2009;22:1303–1311.
Lee WY, Tzeng CC, Chou CY . Uterine leiomyosarcomas coexistent with cellular and atypical leiomyomata in a young woman during the treatment with luteinizing hormone-releasing hormone agonist. Gynecol Oncol 1994;52:74–79.
Kampjarvi K, Makinen N, Kilpivaara O et al. Somatic MED12 mutations in uterine leiomyosarcoma and colorectal cancer. Br J Cancer 2012;107:1761–1765.
Matsubara A, Sekine S, Yoshida M et al. Prevalence of MED12 mutations in uterine and extrauterine smooth muscle tumours. Histopathology 2013;62:657–661.
Ravegnini G, Marino-Enriquez A, Slater J et al. MED12 mutations in leiomyosarcoma and extrauterine leiomyoma. Mod Pathol 2013;26:743–749.
Markowski DN, Huhle S, Nimzyk R et al. MED12 mutations occurring in benign and malignant mammalian smooth muscle tumors. Genes Chromosomes Cancer 2013;52:297–304.
Bell SW, Kempson RL, Hendrickson MR . Problematic uterine smooth muscle neoplasms. A clinicopathologic study of 213 cases. Am J Surg Pathol 1994;18:535–558.
Kim SY, Cordeiro MH, Serna VA et al. Rescue of platinum-damaged oocytes from programmed cell death through inactivation of the p53 family signaling network. Cell Death Differ 2013;20:987–997.
Qiang W, Cahill JM, Liu J et al. Activation of transcription factor Nrf-2 and its downstream targets in response to moloney murine leukemia virus ts1-induced thiol depletion and oxidative stress in astrocytes. J Virol 2004;78:11926–11938.
Hodge JC, Kim TM, Dreyfuss JM et al. Expression profiling of uterine leiomyomata cytogenetic subgroups reveals distinct signatures in matched myometrium: transcriptional profilingof the t(12;14) and evidence in support of predisposing genetic heterogeneity. Hum Mol Genet 2012;21:2312–2329.
Mehine M, Kaasinen E, Makinen N et al. Characterization of uterine leiomyomas by whole-genome sequencing. N Engl J Med 2013;369:43–53.
Farquhar CM, Steiner CA . Hysterectomy rates in the United States 1990-1997. Obstet Gynecol 2002;99:229–234.
Zhao SZ, Wong JM, Arguelles LM . Hospitalization costs associated with leiomyoma. Clin Ther 1999;21:563–575.
Mauskopf J, Flynn M, Thieda P et al. The economic impact of uterine fibroids in the United States: a summary of published estimates. J Womens Health (Larchmt) 2005;14:692–703.
Ligon AH, Morton CC . Leiomyomata: heritability and cytogenetic studies. Hum Reprod Update 2001;7:8–14.
Moore SD, Herrick SR, Ince TA et al. Uterine leiomyomata with t(10;17) disrupt the histone acetyltransferase MORF. Cancer Res 2004;64:5570–5577.
Lehtonen R, Kiuru M, Vanharanta S et al. Biallelic inactivation of fumarate hydratase (FH) occurs in nonsyndromic uterine leiomyomas but is rare in other tumors. Am J Pathol 2004;164:17–22.
Garcia-Torres R, Cruz D, Orozco L et al. Alport syndrome and diffuse leiomyomatosis. Clinical aspects, pathology, molecular biology and extracellular matrix studies. A synthesis, Nephrologie 2000;21:9–12.
Acknowledgements
We acknowledge the Northwestern University Genomic Core Facility; Dongjun Ren from the Richard Miller Lab in the department of Molecular Pharmacology and Biological Chemistry; Jessica Choy and Vanida A Serna from the Kurita lab for technical assistance. This study is supported in part by funds from the Edna Leiomyosarcoma Foundation (JJW), RO1 HD064402 (T.K), and RO1 CA154358 (TK).
Author contributions
Design: JJW, TK. Experiments: EB, WQ, QZ, MEF, SD, YL, JJW. Materials: JJW, TK, BHK. Analysis: JJW, WQ, TK. Manuscript: JJW.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Bertsch, E., Qiang, W., Zhang, Q. et al. MED12 and HMGA2 mutations: two independent genetic events in uterine leiomyoma and leiomyosarcoma. Mod Pathol 27, 1144–1153 (2014). https://doi.org/10.1038/modpathol.2013.243
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2013.243
Keywords
This article is cited by
-
Uterine leiomyoma with RAD51B::NUDT3 fusion: a report of 2 cases
Virchows Archiv (2023)
-
The Mediator Complex Subunit 12 (MED-12) Gene and Uterine Fibroids: a Systematic Review
Reproductive Sciences (2023)
-
Myometrial oxidative stress drives MED12 mutations in leiomyoma
Cell & Bioscience (2022)
-
Tryptophan 2,3-Dioxygenase-2 in Uterine Leiomyoma: Dysregulation by MED12 Mutation Status
Reproductive Sciences (2022)
-
Frequency of MED12 Mutation in Relation to Tumor and Patient’s Clinical Characteristics: a Meta-analysis
Reproductive Sciences (2022)