Abstract
Characterization of tumor genetics and epigenetics allows to stratify a tumor entity according to molecular pathways and may shed light on the interactions of different types of DNA alterations during tumorigenesis. Small intestinal adenocarcinoma is rare, and to date the interrelation of genomic instability and epigenetics has not been investigated in this tumor type. We therefore analyzed 37 primary small bowel carcinomas with known microsatellite instability and KRAS status for chromosomal instability using comparative genomic hybridization, for the presence of aberrant methylation (CpG island methylation phenotype) by methylation-specific polymerase chain reaction, and for BRAF mutations. Chromosomal instability was detected in 22 of 37 (59%) tumors (3 of 9 microsatellite instable, and 19 of 28 microsatellite stable carcinomas). Nine carcinomas (24%) were microsatellite and chromosomally stable. High-level DNA methylation was found in 16% of chromosomal instable tumors and in 44% of both microsatellite instable and microsatellite and chromosomally stable carcinomas. KRAS was mutated in 55, 0, and 10% of chromosomal instable, microsatellite instable, and microsatellite and chromosomally stable tumors, respectively whereas the frequencies of BRAF mutations were 6% for chromosomal instable and 22% for both microsatellite instable and microsatellite and chromosomally stable carcinomas. In conclusion, in this study we show that chromosomal instable carcinomas of the small intestine are distinguished from microsatellite instable and microsatellite and chromosomally stable tumors by a high frequency of KRAS mutations, low frequencies of CpG island methylation phenotype, and BRAF mutations. In microsatellite instable and microsatellite and chromosomally stable cancers, CpG island methylation phenotype and BRAF/KRAS mutations are similarly distributed, indicating common mechanisms of tumor initiation or progression in their molecular pathogenesis.
Similar content being viewed by others
Main
Although the small intestine represents the largest segment of the gastrointestinal tract and is located between two areas of high cancer risk, the stomach and the colon, adenocarcinoma development is exceedingly rare in this location (incidence 1/100 000, SEER Cancer Statistics 2003–20071). Small bowel adenocarcinoma and colorectal cancer show overlaps in morphology and hereditary and non-hereditary risk factors, including the hereditary non-polyposis colorectal cancer syndrome, familial adenomatous polyposis, and Crohn's disease.2, 3, 4 Recent studies, however, provide evidence for a somewhat different molecular pathogenesis of the majority of sporadic carcinomas in both locations.5, 6, 7, 8, 9
Microsatellite and chromosomal instability (MSI and CIN) are two divergent types of genomic instability found in adenocarcinomas throughout the gastrointestinal tract, including the small intestine.6, 7, 9, 10, 11, 12 MSI is caused by inactivation of the DNA mismatch repair system that may either result from promoter hypermethylation of the DNA mismatch repair gene MLH1 in sporadic carcinogenesis,13 or following germline mutations in DNA mismatch repair system genes in hereditary nonpolyposis colorectal cancer.14 CIN involves gains or losses of whole chromosomes and/or chromosomal fractions and is considered to result from defects in chromosomal segregation, telomere stability, and DNA damage repair.15, 16 In ∼80% of colon cancers, CIN and/or MSI are found. A share of 20% of colorectal cancers, however, is microsatellite and chromosomally stable (MACS), and the molecular pathogenesis of these colorectal cancers is not completely understood.17, 18
The CpG island methylation phenotype (CIMP) is an epigenetic phenomenon found in a variety of cancers.19, 20 In colorectal and gastric cancers, CIMP frequently associates with MLH1 methylation, loss of MLH1 transcription, and MSI-H.19, 21, 22 An association of CIMP with BRAF mutations has been found in colorectal cancer23 but, interestingly, mutations in BRAF do not have a significant role in either CIMP-positive or -negative gastric cancers.24
Analyses of genomic and epigenetic interrelations in human neoplasms have added largely to our understanding of tumor development and have led to a novel molecular classification of colorectal cancer.25 As the pathogenesis of small bowel adenocarcinoma is not completely understood and interrelations of epigenetic and genetic alterations have not been investigated in this tumor type, we analyzed 37 primary small bowel adenocarcinomas stratified by MSI and KRAS status for chromosomal imbalances, CIMP, and the presence of BRAF mutations. In this study we show that the majority of small bowel adenocarcinomas display genomic instability (either CIN and/or MSI) and that a subset of 24% of tumors is microsatellite and chromosomally stable. We further demonstrate similar epigenetic alterations and relations of BRAF/KRAS mutations in MSI-H and MACS but not in CIN cancers, indicating MSI-H and MACS carcinogenesis to follow similar initializing or promoting epigenetic and genetic events.
Materials and methods
Tumor Samples
A total of 37 unselected primary adenocarcinomas of the small intestine, including 15 duodenal, 7 jejunal, and 7 ileal tumors, as well as 8 adenocarcinomas in small intestinal segment resection without specification of jejunal or ileal origin, were analyzed. Familial adenomatous polyposis coli, juvenile polyposis, and Peutz–Jeghers-syndrome were excluded using pathologic examination. One carcinoma (884) was associated with celiac disease and one (271) with Crohn's disease. The duodenal cancer (320) was diagnosed in a patient with hereditary nonpolyposis colorectal cancer. Hereditary nonpolyposis colorectal cancer was considered in two other patients with MSI-H carcinomas (884, 235). In one of these patients, germline analysis of the MLH1 gene (884) revealed no mutation. In the second patient, no germline analysis has been performed. Patient age ranged from 29 to 84 years (median age 64). The clinical data are given in Table 1. The study was approved by the institutional ethics committee (application no. 206/05) at the Medical Faculty at Heidelberg University.
DNA Isolation
For the isolation of DNA from tumor samples for methylation-specific PCR, as well as for mutational analysis of BRAF, tissue slides of 5-μm thickness were prepared. Tumor tissue was isolated under microscopic control of unstained slides by manual microdissection. For comparative genomic hybridization (CGH), areas of high tumor cell concentration (>70%) were marked on the slide and the corresponding area was removed from the tumor block with the help of a 0.2 mm punch needle. For CGH and PCR analyses, DNA was prepared using the DNeasy tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
MSI Analysis
Tumor samples had previously been typed for MSI as described.26 In brief, DNA isolated from the tumors and non-neoplastic normal mucosa was amplified. MSI analysis was performed using the standard NCI/ICG-hereditary nonpolyposis colorectal cancer marker panel27 and CAT25.28 PCR products were separated on an ABI3700 automated sequencer (Applied Biosystems, Warrington, UK). Tumors were classified as MSI-H if at least two of the markers displayed a shift in product length compared with the product of non-neoplastic mucosa.
CGH Analysis
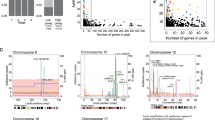
CGH was performed as previously described in detail.29 In brief, tumor DNA was biotinylated and normal control DNA was extracted from healthy human placenta tissue and labeled with digoxigenin. After hybridization of the labeled DNA on metaphase spreads (CGH target slides; Abbott Molecular, Abbott Park, IL, USA) at 37 °C for 48 h, the slides were incubated with fluorescence-labeled anti-digoxigenin and anti-biotin antibodies. Metaphase spreads from each specimen were analyzed and photodocumented using an Axiovert S100 microscope (Zeiss, Jena, Germany), a CCD camera, and hardware and software as supplied by Metasystems (Altlussheim, Germany). Fluorescence ratios were determined using both fixed cutoff values (0.8 for losses and 1.25 for gains) and cutoff values with a twofold s.d. Chromosomal abnormalities detected with both approaches were scored. Centromeric and telomeric chromosomal areas as well as chromosome 19 were excluded from scoring. CGH data of 21 of the 37 carcinomas have previously been reported.7, 8
Methylation-Specific PCR
Methylation-specific PCR was performed for five CpG islands known to be methylated in colorectal cancer: MINT1, MINT2, MGMT, MLH1, and p16 (CDKN2A). Primers and PCR conditions were used as described previously (MINT1 and MINT2;30 MGMT;31 p16;32 MLH133). DNA was bisulfite converted using the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA) according to the manufacturer's instructions. One-fifth of a conversion reaction was used as template.
PCR products were visualized on 2% agarose TAE gel or 6% nondenaturing polyacrylamide gels (PAA). Methylation status was assessed based on the presence or absence of amplified product in the methylation-specific PCR. CIMP-high (CIMP-H), CIMP-low (CIMP-L), and CIMP-negative (CIMP-neg) were scored when >50, 20–50%, and 0% of the investigated loci were positive for methylation-specific PCR, respectively (Figure 1). Methylation-specific PCR data of 21 carcinomas were the subject of a previous study on the pathogenesis of celiac disease-associated small bowel adenocarcinomas.34
BRAF Mutations
For analysis of BRAF mutations, the following primers were used: BRAF forward 5′-CCTAAACTCTTCATAATGCTTGCTC-3′, BRAF reverse 5′-CCACAAAATGGATCCAGACA-3′. PCR conditions were as follows: denaturing 92 °C (45 s), annealing at 60 °C (1 min), extension 72 °C (1 min); 35 cycles. PCR products were analyzed by single-strand conformation polymorphism (SSCP) method. Tumors with aberrantly shifted bands were subjected to sequencing. The PCR fragments of these tumors were purified using the high Pure PCR Purification kit (Roche Diagnostics, Mannheim, Germany) as recommended by the manufacturer, subjected to direct cycle sequencing using a bigDye termination kit (Applied Biosystems), and automated sequencing by ABIPrism 377 (Applied Biosystems).
Results
Stratification of Small Bowel Adenocarcinomas According to Chromosomal and Microsatellite Instability Identifies Four Genomic Subgroups
Of 37 carcinomas, 22 (19 microsatellite stable, 3 MSI-H) displayed chromosomal imbalances. According to the microsatellite status and the presence of chromosomal imbalances, four groups of tumors could be distinguished.
Group 1, MSS/CIN
In all, 19 microsatellite stable (MSS) carcinomas displayed chromosomal imbalances. The most consistent finding was a deletion at chromosome 18q found in 11 of 19 carcinomas in this group. This alteration was commonly found in combination with additional chromosomal imbalances. Deletions at a lower frequency were detected for chromosomal arms 4q (5/19), 6q (4/19), 8p (4/19), and 17p (4/19) whereas gains were common at chromosomes 20q (8/19) and chromosome 7 (5/19).
Group 2, MSI-H/CIN
In all, three MSI-H carcinomas displayed chromosomal alterations. Two tumors showed gains at chromosome 8. No other recurrent imbalances were found. All tumors lacked deletions at 18q.
Group 3, MSI-H/CIN negative
Of nine MSI-H tumors, six did not show chromosomal alterations by CGH analysis.
Group 4, MSS/CIN negative
Of 28 MSS tumors, 9 did not show chromosomal alterations by CGH analysis. The results of CGH are summarized in Table 1.
Stratification of Small Bowel Adenocarcinomas According to Methylation Status: CIMP-H Is Associated with MSI-H and MACS
Among the 37 investigated carcinomas, 24 tumors were found to be methylated for at least one investigated locus. A total of 11 carcinomas were scored CIMP-H (3 CIN/MSS, 4 MSI-H, and 4 MACS), 13 carcinomas CIMP-L (4 MSI-H, 6 CIN/MSS, and 3 MACS), and 13 CIMP-neg (10 CIN/MSS, 1 MSI-H, and 2 MACS). CIMP-H was found in 15% (3/19) of CIN/MSS carcinomas, in 44% (4/9) of MSI-H, and in 44% (4/9) of MACS tumors. MINT2 and MINT1 represented the most commonly methylated loci (17 and 16 cases, respectively), followed by methylation at MGMT (14 cases), p16 (8 cases), and MLH1 (7 MSI-H and 1 MSS case). Whereas methylations at single loci were found for MGMT, MINT1, and MINT2, methylation at MLH1 was only seen in combination with at least one additionally methylated locus, and methylation of p16 was only observed in CIMP-H carcinomas.
BRAF Mutations Are Associated with MSI-H, MACS, and CIMP
BRAF mutations were observed in five carcinomas. In four cases, a codon 600 mutation (GTG to GAG; V600E) was found, and one additional case displayed a previously published mutation consisting of a 3-bp deletion (c1869–187135) predicted to result in a loss of W604 on the protein level. All tumors with BRAF mutations were CIMP positive, including three CIMP-H and two CIMP-L carcinomas. BRAF mutations were observed in 2 of 9 MSI-H carcinomas, in 2 of 9 MACS carcinomas, and in 1 of 19 CIN carcinomas.
Discussion
In this study, we aimed at elucidating the association of genetics, epigenetics, and mutations in KRAS/BRAF in small bowel adenocarcinomas. As a first step, we therefore stratified 37 carcinomas by the presence of genomic instability. Among 9 MSI-H and 28 MSS carcinomas investigated, we identified 3 of the MSI-H (33%) and 19 of the MSS carcinomas (68%) to display CIN. We thus found MSI-H to be inversely associated with CIN. Moreover, compared with MSS cancers with CIN, the extents and chromosomal locations of CIN in MSI-H cancers were different (Table 1), indicating a selective growth advantage of divergent chromosomal alterations in MSI-H and MSS cancers. These data are comparable with the situation in colorectal cancer, in which generally lower numbers and different types of chromosomal alterations are detected in MSI-H compared with MSS carcinomas.36, 37
MSS small bowel carcinomas with CIN presented the largest genetic subgroup (51%). Among the alterations seen in this group, deletions at chromosome 18q were the most common shared feature (58% in the CIN group, 30% of all investigated carcinomas) and were commonly associated with gains at chromosomes 20q and 7 as well as with losses of chromosomes 4q, 8p, and 17p (Table 1). Our finding of 30% 18q deletions among all investigated small bowel adenocarcinomas is in contrast with a previous matrix CGH study by Diosdado et al38 reporting <10% of 18q deletions in 48 small intestinal adenocarcinoma, whereas the frequencies of deletions at 4q, 8p, 17p, and gains at chromosomes 7 and 20 were similar to those of our study. The reason for this discrepancy is not clear. It may be explained by sampling variation, and in part result from the high number of celiac disease-associated carcinomas (15/48) included in the study by Diosdado et al38 (1/37 in our study), which have been shown to follow a somewhat different genetic pathway than nonceliac disease-related sporadic carcinomas.38, 39
Chromosome 18q harbors the SMAD4 gene, a mediator of TGFβ signaling, which was previously described as a mutational target for a variety of cancers, including small bowel adenocarcinoma.7, 40 The importance of SMAD4 inactivation for small bowel carcinogenesis is underlined by the fact that deletions at the SMAD4 locus were found in two carcinomas with only minimal numbers of other chromosomal imbalances (T401, T970), and that somatic intragenic mutations of SMAD4 were identified in two tumors lacking 18q loss (T246, T502; data not shown).
Although the majority of the carcinomas investigated in this study could be assigned to one type of genomic instability, a significant share of 24% of tumors were found microsatellite and chromosomally stable. This finding is similar to the situation in colorectal cancer, where MACS is identified in 15–30% of tumors.17, 41, 42, 43 The pathogenesis of MACS tumors is not understood. It is noteworthy, however, that we have previously identified44 large-scale N-terminal deletion mutations of β-catenin in three small bowel adenocarcinomas characterized as MACS in the present study, indicating that inactivation of DNA repair mechanisms other than those implicated in mismatch or chromosomal repair could be involved in MACS tumorigenesis.
To investigate the interaction of genomic instability types with epigenetics, we analyzed the tumors for CIMP. We observed 11 carcinomas displaying high-level CIMP (CIMP-H). These 11 carcinomas were not evenly distributed among CIN, MSI-H, and MACS cancers. Similar to the situation in colorectal cancer, CIMP-H with MLH1 methylation was particularly frequent in MSI-H carcinomas, providing evidence for a link of CIMP-H and MSI-H in small bowel adenocarcinomas also. Interestingly, we found that CIMP-H is similarly frequent in MACS and is less frequently present in MSS tumors with CIN. These data are in line with a previous observation in colorectal cancer reporting an inverse correlation of aberrant methylation and CIN.45, 46 BRAF mutations have been reported to associate with CIMP-H in colorectal cancer. Our data show that the situation is similar in small bowel cancer. We found BRAF mutations in three CIMP-H and two CIMP-L carcinomas. The BRAF mutation frequency was identical in MSI-H and MACS carcinomas (2/9 cases each), whereas only 1 of 19 CIN carcinomas was BRAF mutated, indicating that not only CIMP-H but also BRAF mutations preferentially occur in the absence of CIN. In contrast, mutations in KRAS were commonly found in MSS/CIN carcinomas (58%) and were infrequent in MSI-H and MACS carcinomas.
Our comprehensive analyses of genetic and epigenetic alterations show that small bowel adenocarcinomas can be stratified into different molecular subgroups. Our findings reveal similar principles of molecular tumorigenesis than recently reported in two comparable studies on colorectal cancer45, 46 (Figure 2), and indicate that CIMP is related to MSI- and MACS-associated intestinal carcinogenesis, whereas it is inversely correlated with CIN. According to our analysis, 50% of small bowel adenocarcinomas are CIN, frequently KRAS mutated, and usually show no or a low level of methylation. Among the remaining 50% of chromosomally stable tumors, MSI-H and MACS cancers had an equal share. MSI-H and MACS small bowel carcinomas usually lack KRAS mutations and display similar frequencies of high-level methylation and BRAF mutations, indicating common mechanisms to trigger MSI-H and MACS. Future analysis may show if the molecular subtypes of small bowel adenocarcinoma correlate with tumor biology and prognosis.
Comparison of the distribution of CIN, MSI, and MACS in small intestinal adenocarcinoma and colorectal cancer. The results of this study and the average values of two similar studies on colorectal cancer45, 46 are presented. The frequency of CIMP-H in the individual genetic subtypes highlights the predominant occurrence of CIMP-H in MSI and MACS carcinomas. *The frequency of CIMP-H in MSI/CIN colorectal carcinomas could only be calculated for one study45 (0/3 cases).
References
Schottenfeld D, Beebe-Dimmer JL, Vigneau FD . The epidemiology and pathogenesis of neoplasia in the small intestine. Ann Epidemiol 2009;19:58–69.
Watson P, Lynch HT . Extracolonic cancer in hereditary nonpolyposis colorectal cancer. Cancer 1993;71:677–685.
Fearnhead NS, Britton MP, Bodmer WF . The ABC of APC. Hum Mol Genet 2001;10:721–733.
Piton G, Cosnes J, Monnet E, et al. Risk factors associated with small bowel adenocarcinoma in Crohn's disease: a case-control study. Am J Gastroenterol 2008;103:1730–1736.
Arai M, Shimizu S, Imai Y, et al. Mutations of the Ki-ras, p53 and APC genes in adenocarcinomas of the human small intestine. Int J Cancer 1997;70:390–395.
Rashid A, Hamilton SR . Genetic alterations in sporadic and Crohn's-associated adenocarcinomas of the small intestine. Gastroenterology 1997;113:127–135.
Blaker H, von Herbay A, Penzel R, et al. Genetics of adenocarcinomas of the small intestine: frequent deletions at chromosome 18q and mutations of the SMAD4 gene. Oncogene 2002;21:158–164.
Blaker H, Mechtersheimer G, Sutter C, et al. Recurrent deletions at 6q in early age of onset non-HNPCC- and non-FAP-associated intestinal carcinomas. Evidence for a novel cancer susceptibility locus at 6q14-q22. Genes Chromosomes Cancer 2008;47:159–164.
Wheeler JM, Warren BF, Mortensen NJ, et al. An insight into the genetic pathway of adenocarcinoma of the small intestine. Gut 2002;50:218–223.
Planck M, Ericson K, Piotrowska Z, et al. Microsatellite instability and expression of MLH1 and MSH2 in carcinomas of the small intestine. Cancer 2003;97:1551–1557.
Hibi K, Kondo K, Akiyama S, et al. Frequent genetic instability in small intestinal carcinomas. Jpn J Cancer Res 1995;86:357–360.
Brueckl WM, Heinze E, Milsmann C, et al. Prognostic significance of microsatellite instability in curatively resected adenocarcinoma of the small intestine. Cancer Lett 2004;203:181–190.
Cunningham JM, Christensen ER, Tester DJ, et al. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res 1998;58:3455–3460.
Lynch HT, de la Chapelle A . Hereditary colorectal cancer. N Engl J Med 2003;348:919–932.
Gollin SM . Mechanisms leading to chromosomal instability. Semin Cancer Biol 2005;15:33–42.
Geigl JB, Obenauf AC, Schwarzbraun T, et al. Defining ‘chromosomal instability’. Trends Genet 2008;24:64–69.
Tang R, Changchien CR, Wu MC, et al. Colorectal cancer without high microsatellite instability and chromosomal instability—an alternative genetic pathway to human colorectal cancer. Carcinogenesis 2004;25:841–846.
Ostwald C, Linnebacher M, Weirich V, et al. Chromosomally and microsatellite stable colorectal carcinomas without the CpG island methylator phenotype in a molecular classification. Int J Oncol 2009;35:321–327.
Bariol C, Suter C, Cheong K, et al. The relationship between hypomethylation and CpG island methylation in colorectal neoplasia. Am J Pathol 2003;162:1361–1371.
Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 1999;96:8681–8686.
Kim H, Kim YH, Kim SE, et al. Concerted promoter hypermethylation of hMLH1, p16INK4A, and E-cadherin in gastric carcinomas with microsatellite instability. J Pathol 2003;200:23–31.
Fleisher AS, Esteller M, Wang S, et al. Hypermethylation of the hMLH1 gene promoter in human gastric cancers with microsatellite instability. Cancer Res 1999;59:1090–1095.
Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006;38:787–793.
Wu M, Semba S, Oue N, et al. BRAF/K-ras mutation, microsatellite instability, and promoter hypermethylation of hMLH1/MGMT in human gastric carcinomas. Gastric Cancer 2004;7:246–253.
Jass JR . Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 2007;50:113–130.
Michel S, Kloor M, Singh S, et al. Coding microsatellite instability analysis in microsatellite unstable small intestinal adenocarcinomas identifies MARCKS as a common target of inactivation. Mol Carcinog 2010;49:175–182.
Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248–5257.
Findeisen P, Kloor M, Merx S, et al. T25 repeat in the 3′ untranslated region of the CASP2 gene: a sensitive and specific marker for microsatellite instability in colorectal cancer. Cancer Res 2005;65:8072–8078.
Isola J, DeVries S, Chu L, et al. Analysis of changes in DNA sequence copy number by comparative genomic hybridization in archival paraffin-embedded tumor samples. Am J Pathol 1994;145:1301–1308.
Chan AO, Issa JP, Morris JS, et al. Concordant CpG island methylation in hyperplastic polyposis. Am J Pathol 2002;160:529–536.
Esteller M, Hamilton SR, Burger PC, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 1999;59:793–797.
Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996;93:9821–9826.
Park SJ, Rashid A, Lee JH, et al. Frequent CpG island methylation in serrated adenomas of the colorectum. Am J Pathol 2003;162:815–822.
Bergmann F, Singh S, Michel S, et al. Small bowel adenocarcinomas in celiac disease follow the CIM-MSI pathway. Oncol Rep 2010;24:1535–1539.
Blaker H, Helmchen B, Bonisch A, et al. Mutational activation of the RAS-RAF-MAPK and the Wnt pathway in small intestinal adenocarcinomas. Scand J Gastroenterol 2004;39:748–753.
Li LS, Kim NG, Kim SH, et al. Chromosomal imbalances in the colorectal carcinomas with microsatellite instability. Am J Pathol 2003;163:1429–1436.
Trautmann K, Terdiman JP, French AJ, et al. Chromosomal instability in microsatellite-unstable and stable colon cancer. Clin Cancer Res 2006;12:6379–6385.
Diosdado B, Buffart TE, Watkins R, et al. High-resolution array comparative genomic hybridization in sporadic and celiac disease-related small bowel adenocarcinomas. Clin Cancer Res 2010;16:1391–1401.
Potter DD, Murray JA, Donohue JH, et al. The role of defective mismatch repair in small bowel adenocarcinoma in celiac disease. Cancer Res 2004;64:7073–7077.
Svrcek M, Jourdan F, Sebbagh N, et al. Immunohistochemical analysis of adenocarcinoma of the small intestine: a tissue microarray study. J Clin Pathol 2003;56:898–903.
Hawkins NJ, Tomlinson I, Meagher A, et al. Microsatellite-stable diploid carcinoma: a biologically distinct and aggressive subset of sporadic colorectal cancer. Br J Cancer 2001;84:232–236.
Yao J, Eu KW, Seow-Choen F, et al. Microsatellite instability and aneuploidy rate in young colorectal-cancer patients do not differ significantly from those in older patients. Int J Cancer 1999;80:667–670.
Chan TL, Curtis LC, Leung SY, et al. Early-onset colorectal cancer with stable microsatellite DNA and near-diploid chromosomes. Oncogene 2001;20:4871–4876.
Breuhahn K, Singh S, Schirmacher P, et al. Large-scale N-terminal deletions but not point mutations stabilize beta-catenin in small bowel carcinomas, suggesting divergent molecular pathways of small and large intestinal carcinogenesis. J Pathol 2008;215:300–307.
Cheng YW, Pincas H, Bacolod MD, et al. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin Cancer Res 2008;14:6005–6013.
Goel A, Nagasaka T, Arnold CN, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology 2007;132:127–138.
Acknowledgements
We gratefully acknowledge Stefanie Kellner for perfect technical assistance. Funding was by DFG (Bl 554–3/2).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Warth, A., Kloor, M., Schirmacher, P. et al. Genetics and epigenetics of small bowel adenocarcinoma: the interactions of CIN, MSI, and CIMP. Mod Pathol 24, 564–570 (2011). https://doi.org/10.1038/modpathol.2010.223
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2010.223
Keywords
This article is cited by
-
Clinical practice guidelines for duodenal cancer 2021
Journal of Gastroenterology (2022)
-
Smoking, alcohol consumption and colorectal cancer risk by molecular pathological subtypes and pathways
British Journal of Cancer (2020)
-
Trends in incidence of small bowel cancer according to histology: a population-based study
Journal of Gastroenterology (2020)
-
A prognostic CpG score derived from epigenome-wide profiling of tumor tissue was independently associated with colorectal cancer survival
Clinical Epigenetics (2019)
-
Molecular alterations and PD-L1 expression in non-ampullary duodenal adenocarcinoma: Associations among clinicopathological, immunophenotypic and molecular features
Scientific Reports (2019)