Abstract
Despite the high number of previous studies, the role of p53 alterations in prostate cancer is not clearly defined. To address the role of p53 alterations in prostate cancer biology, a total of 2514 cancers treated by radical prostatectomy were successfully analyzed by immunohistochemistry in a tissue microarray format. Overall a low rate of p53-positive tumors was found (2.5%). A significant underestimation of p53-positive cases was excluded by subsequent large section analyses and direct sequencing of the p53 gene in subsets of our patients. Large section analysis of 23 cases considered negative on the tissue microarray yielded only one weakly p53-positive tumor. Only 4 out of 64 (6.4%) high-grade tumors, that were considered negative for p53 by immunohistochemistry, presented exon 5–8 mutations. These data suggest a high sensitivity of our immunohistochemistry approach and confirm the overall low frequency of p53 alterations in clinically localized prostate cancer. A positive p53 immunostaining was strongly associated with presence of exon 5–8 mutations (P<0.0001), advanced pT-stage (P<0.0001), high Gleason grade (P<0.0001), positive surgical margins (P=0.03) and early biochemical tumor recurrence (P<0.0001). A higher rate of positive p53 immunostaining was detected in late-stage diseases including metastatic prostate cancer (P=0.0152) and hormone-refractory tumors (P=0.0003). Moreover, p53 expression was identified as an independent predictor of biochemical tumor recurrence in the subgroup of low- and intermediate-grade cancers. In summary, the results of this study show that p53 mutations characterize a small biologically aggressive subgroup of prostate cancers with a high risk of progression after prostatectomy. The rate of p53 alterations increases with prostate cancer progression.
Similar content being viewed by others
Main
Inactivation of the p53 tumor suppressor is one of the most frequent genetic alterations in malignant tumors. p53 inactivation derails cellular programs inducing apoptosis in DNA damaged cells and consequently enables tumor progression through acquisition of additional genetic changes.1 In most cases, p53 inactivation is partly due to an inactivating mutation of one p53 allele. As many of these mutations lead to a prolonged half-life of p53 protein, immunohistochemistry is commonly used to detect p53-inactivated cancers. A high number of immunohistochemistry studies have been conducted to investigate the role of p53 inactivation in various cancer types. Several of them suggested a link between nuclear p53 protein accumulation and poor prognosis. However, these results were not confirmed by other studies.2 For this reason, p53 testing has not become a routine procedure in the evaluation of any of these tumors.2 In prostate cancer, the clinical relevance of p53 alterations is unclear. p53 alterations were analyzed in almost 1000 studies. Most of them suggest that immunohistochemical p53 positivity increases with high grade, advanced stage and peripheral zone origin.3, 4, 5, 6, 7 Some studies have suggested that nuclear p53 accumulation may correlate with poor prognosis after radical prostatectomy,8, 9 external beam radiation,10 and watchful waiting11 but these data were not confirmed in other studies.12, 13, 14 Perhaps some of these discordances were caused by the relatively small number of patients included in these studies ranging from 24–392 patients.6, 15
Despite the high number of previous studies, the role of p53 alterations in prostate cancer and other malignancies is not clearly defined. The range of p53-positive prostate cancers reported in the literature ranges from 4 to 61%.6, 16 Some of these discrepancies could theoretically be attributed to differences in the examined cohorts with variable fractions of high-risk tumors. It seems more likely, however, that most controversial p53 immunohistochemistry results are caused by technical issues such as the selected reagents and protocols. Considering that p53 protein is physiologically expressed in activated cell nuclei, a too sensitive detection system can easily identify positive p53 staining in cancers or normal tissues without p53 mutations.
In order to clarify epidemiology and prognostic significance of p53 alterations in prostate cancer we conducted a large-scale study involving more than 2500 prostate cancers homogeneously treated in our center. A tissue microarray format was utilized allowing the simultaneous immunohistochemical analysis of all tumors in one day with one set of reagents thus enabling maximal experimental standardization. The utilized immunohistochemistry protocol was validated by comparative sequencing of the p53 gene in more than 100 cases. The results reveal that p53 alterations are infrequent in primary prostate cancer but have high prognostic relevance.
Materials and methods
Patients
Radical prostatectomy specimens were available from 3261 patients, treated at the Department of Urology, University Medical Center, Hamburg-Eppendorf between 1992 and 2005 (Table 1). Follow-up data were available for 2385 patients, ranging from 1 to 144 months (mean, 34 months). None of the patients received adjuvant therapy. Additional (salvage) therapy was only initiated in case of a biochemical relapse. All prostatectomy specimens were analyzed according to a standard procedure. All prostates were completely paraffin-embedded, including whole-mount sections as previously described.3 All hematoxylin and eosin-stained histological sections from all prostatectomy specimens were reviewed for the purpose of this study and the index tumors, as defined by the largest tumor focus and/or the focus with the worst Gleason pattern, were marked on the slides. One 0.6 mm tissue core was punched out from the index tumors of each case, and transferred in a tissue microarray format as described.17 The 3261 cores were distributed among seven tissue microarray blocks, each containing 129–522 tumor samples. Each tissue microarray block also contained various control tissues including normal prostate tissue, other normal tissues and a set of tumor tissues including several colon and breast cancers with abnormal p53 status (positive controls for immunohistochemistry). In addition, 37 lymph node metastases and 35 hormone-refractory cancers were analyzed on a separate prognosis-tissue microarray. Hormone-refractory prostate cancer was defined as serum castration levels of testosterone, three consecutive rises of the prostatic specific antigene (PSA) resulting in two 50% increases over the nadir, anti-androgen withdrawal for at least 4 weeks, PSA progression despite secondary hormonal manipulations, or progression of osseous or soft tissue lesions.18 To address the role of the potential impact of tumor heterogeneity, 28 selected tumors (5 positive, 23 negative) were additionally analyzed on large sections.
Immunohistochemistry
Freshly cut tissue microarray sections were analyzed in one day in one experiment for each antibody. Slides were immunostained for low molecular weight cytokeratins to assure presence of cancer cells in the tissue microarray spots. For this purpose, the antibody 34βE12 (clone MA903; Dako; 1:12.5) was used for basal cell detection after boiling the sections in an autoclave in citrate buffer, pH 7.8. The antibody DO1 (Oncogene; 1:3600) was used for p53 protein detection at identical pretreatment conditions. The Envision system (DAKO) was used for both antibodies to visualize the immunostainings. Colon cancers with known p53 alterations served as positive controls and normal prostate tissue as negative controls on each tissue microarray section. Only tissue samples with distinct loss of basal cells (proven prostate cancers) were used for p53 analysis. In these samples, p53 positivity was assumed if more than 1% of tumor cells showed unequivocal nuclear staining. For comparison between p53 mutations and p53 expression the staining intensity was qualified in a 5-step scale (0–4). To demonstrate the impact of immunohistochemistry protocol modifications on p53 data in prostate cancer, tissue microarray sections were also analyzed using DO1 at a dilution of 1:20.
p53 Sequencing
All 62 cancers with detectable p53 expression and 63 p53 negative high-grade tumors (Gleason score: ≥4+3) were sequenced for p53 exon 5–8 mutations. In one p53-positive case, DNA was not suitable for PCR amplification. DNA was extracted from a separate tissue core taken from a tumor area adjacent to the core used for tissue microarray construction. The QIAamp DNA mini kit (Qiagen, Hilden, Germany) was utilized according to the manufacturer's instructions. Isolated DNA (20–300 ng) were used as template for amplification of the p53 exons 5–8 using the following primers (given in 5′ to 3′ direction)—exon 5 forward: CACTTGTGCCCTGACTTTCAAC, exon 5 reverse: CAACCAGCCCTGTCGTCTCTC (product length 268 bp); exon 6 forward: TCCCCAGGCCTCTGATTCCT, exon 6 reverse: CCTTAACCCCTCCTCCCAGA (product length 190 bp); exon 7 forward: GCCTCATCTTGGGCCTGTGTTATC, exon 7 reverse: TCAGAGGCAAGCAGAGGCTG (product length 203 bp); exon 8 forward: CTGATTTCCTTACTGCCTCTTGC, exon 8 reverse: TCTCCTCCACCGCTTCTTGTC q(product length 216 bp).
Amplification took place in 25 μl reactions with 50 cycles each of 10 s at 95°C, 20 s at 55°C and 40 s at 72°C. After ethanol precipitation of the PCR products, the sequencing reactions in both directions were performed using the same primers and the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Weiterstadt, Germany). Sequence analysis was carried out on an ABI PRISM 3100 genetic analyzer (Applied Biosystems). All sequence variations were confirmed in a second PCR-sequencing reaction.
Statistics
Statistical calculations were performed with PRISM 2.01 software (GraphPad, San Diego, USA). Contingency tables were calculated with the χ2-test and Fisher's exact test. Survival curves were calculated by the Kaplan–Meier method and compared with the Logrank test. Cox regression was used to assess independence of preoperative parameters and p53 expression to predict PSA recurrence after radical prostatectomy.
Technical Issues
Unequivocal prostate cancer was present in 2514 of 3261 arrayed tissue samples. Noninformative cases were caused by missing spots on the tissue microarray (129; 4%) or absence of definite invasive cancer tissue in the associated 34βE12 immunohistochemistry (618; 19%). The latter spots contained normal prostatic tissue, high-grade prostatic intraepithelial neoplasia, or stromal tissue only.
Results
p53 Immunohistochemistry
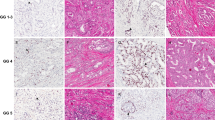
Using our standard p53 protocol (1:3600), a positive p53 staining could be observed in 62 of 2514 cancers (2.5%). Representative images are given in Figure 1. p53 positivity was significantly more frequent in non-organ-confined tumors (P<0.001) and in tumors with higher Gleason grades (P<0.001) or positive surgical margins (P=0.094; Table 2). On a separate progression-tissue microarray the rate of p53-positive cases was higher among 37 interpretable metastases (16.2%; P=0.0152 for metastases vs primary tumors) and 35 hormone-refractory cancers (25.7%, P=0.0003 for hormone-refractory cancers vs primary tumors; Table 3). The relationship with large section p53 immunostaining in 28 selected cases (5 positive on tissue microarray, 23 negative on tissue microarray) showed a concordance of the data in 25 of 26 cases (96%). Only one tissue microarray negative cancer showed a very mild (20%/1+) staining on the corresponding large section. However, a clearly heterogeneous p53 immunostaining was found in two of the five p53-positive large sections. An example is given in Figure 1c. As expected, our modified ‘oversensitive’ p53 protocol (1:20 dilution of the antibody) yielded completely different results. Here more than 90% of all tissue microarray samples showed moderate to strong p53 immunostaining (Figure 2).
p53 Sequence Analysis
Mutations were found in 29 of 63 analyzed p53-positive tumors (46%) by immunohistochemistry. These mutations most frequently occurred in exon 7 (11 cases) and exon 8 (9 cases). Remarkably, one tumor had mutations in both exons 7 and 8. All mutations are described in Table 4. Most mutations were missense mutations but there were also two tumors with a frameshift caused by small deletions. One of these tumors displayed a deletion of 33 base-pairs (bp) in exon 7 and the other tumor had a 17 bp deletion including 8 bp from introns 7–8 and 9 bp from exon 8. Mutations were significantly less frequent in the group of selected high-grade p53 negative (by immunohistochemistry) cancers. Here, only 4 of 63 cases (6.4%) had detectable mutations (P<0.0001 positive tumors vs negative high-grade cancers). At least one of 4 detected mutations in the immunohistochemistry-negative group resulted in a stop codon and consecutively a truncated protein (Table 4). Accordingly a ‘false’ negative immunohistochemistry result had to be expected in this case. Another case in this group displayed a splice mutation in the region at exon 6/introns 6–7. It could be speculated that this mutation might have led to a severely altered protein being undetectable by immunohistochemistry, too.
Relationship with PSA Recurrence
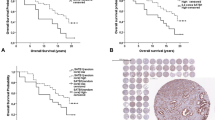
A total of 2385 patients with follow-up data were included into this analysis. The validity of the clinical data attached to the arrayed tissue samples is demonstrated in a first analysis comparing standard clinicopathological parameters (Gleason grade, pT-stage, preoperative PSA serum level, presence of positive surgical margins) with PSA recurrence. All expected associations were found at a high level of statistical significance (P<0.0001 each; Figure 3). Likewise, p53 positivity was significantly related to PSA recurrence. In the p53-positive group, 47.5% of patients presented a biochemical tumor recurrence, as defined by a persisting or rising postoperative PSA (>0.1 ng/ml). In the p53 negative group, biochemical recurrence was detected in only 23.3% of patients. This difference was highly significant (P<0.0001; Figure 4a). Most remarkably, there was no difference in the clinical outcome between p53 immunohistochemistry positive tumors with and without exon 5–8 mutations. A multivariate analysis including preoperative PSA, pT-stage, Gleason grade and surgical margin status demonstrated that p53 immunostaining was an independent predictor of biochemical recurrence (Table 5). More importantly, it could be demonstrated that p53 alteration is an independent predictor of PSA recurrence in low- and intermediate-grade cancer (Gleason ≤3+4; Figure 4b).
Discussion
Only 2.5% of primary prostate cancers were p53 positive by immunohistochemistry. This is lower than those in many previous immunohistochemistry studies where the frequency of p53-positive cases ranged up to 61%.16 It is noteworthy, that the low frequency of p53 alterations detected in this study is unlikely to be caused by a large fraction of false negative results due to our tissue-microarray analysis of only one 0.6 mm tissue sample per prostate cancer. Our comparative large section analysis of 26 cases identified 2 cases with heterogeneous p53 positivity, however, these cases had been identified as p53 positive on tissue microarray. There was only 1 case with mild p53 immunostaining (1+ in 20% of cells) in 23 randomly selected negative cases from the tissue microarray suggesting that we have not missed a significant fraction of p53 altered cases with our approach. Moreover, our sequencing effort involving more than 100 cancers validated our immunohistochemical approach. p53 mutations that were not detected by our immunohistochemistry procedure were only seen in 6.25% of sequenced high-grade cancers (P<0.0001). That at least one of them was probably not detectable by immunohistochemistry due to truncated p53 proteins further argues for the adequacy of our immunohistochemistry procedure. We therefore assume, that the true number of p53-altered cancers was not markedly underestimated for our patient cohort.
A high fraction of organ-confined tumors (pT2) with only 1% p53 positivity in our patient cohort may have contributed to the low overall frequency of p53 alterations, but the increase in our pT3/pT4 group (5% p53 positivity) was not massive. The results obtained with the same protocol on our progression-tissue microarray including metastases (16.2% positive) and hormone-refractory cancers (25.7% positive) demonstrate, however, that the rate of p53 alterations increases further in late stage disease and higher degree of differentiation. Functional studies also suggest a direct correlation of p53 mutations with the transition of prostate cancer to metastatic and hormone-refractory disease.19, 20 Furthermore, p53 expression proved to be an independent predictor of PSA recurrence in the clinically uncertain group of low- and intermediate-grade cancers (Gleason ≤3+4; Figure 3a). These results are in line with other large-scale studies,21 and particularly in this group of cancers a valid biomarker is required to distinguish an aggressive clinical course. However, the drawback of this is that p53 alterations are very rare in low- and intermediate-grade cancers and could therefore be only helpful for a better stratification of a small subset of patients. This will limit the impact of p53 testing in the clinical routine.
Previous studies using prostate cancer tissue microarrays had suggested the use of multiple cores per cancer specimen.22 These recommendations were based on a better concordance of large section findings with tissue microarrays data, if 3–4 cores were utilized per cancer as compared to the use of only one sample. The results of our large-section analysis do not support this notion. It is important to understand, that the selected immunostaining protocol conditions are of such utmost importance for the outcome of immunohistochemistry studies, that the amount of analyzed tissue becomes a parameter of secondary relevance.23 The results obtained with a protocol that was deliberately designed to be ‘oversensitive’ clearly demonstrate that a high rate of positive immunohistochemistry cannot therefore automatically be viewed as an argument for good representativity. A high rate of positive cases can also reflect artificial ‘false’ positivity. It is also possible that alterations of other proteins—such as PTEN inactivation—can lead to a slight p53 protein expression increase in some cases.24 Immunohistochemical p53 mutation analysis is technically challenging because overly sensitive assays can detect physiological p53 expression and therefore lead to false-positive results. The quality of p53 immunohistochemistry results can best be evaluated by comparison with DNA sequencing or clinical outcome data. In prostate cancer, there are no previous tissue-microarray studies comparing results obtained on 1–4 cores with DNA sequencing or clinical outcome data. However, in a series of >500 breast cancers, Torhorst et al25 showed strong associations of p53 positivity with poor survival in 4 different tissue microarrays, each containing one tissue sample of 0.6 mm per tumor. Remarkably, a prognostic significance of p53 immunostaining could not be observed in corresponding large sections, despite 80% more positive cases in this study.25 The most likely explanation of these surprising data is that a dilution of true p53-positive cases by artificial positivities precluded detection of significant associations with survival in the large section analysis.
In this study, although only 38% of our immunohistochemistry positive cancers were confirmed as mutated by direct sequencing of the p53 gene, we do not assume that the number of false-positive immunostainings was unacceptably high. This is especially supported by the similar prognosis of p53 mutated and unmutated immunohistochemistry-positive cancers. We rather assume that some mutations were missed because they were located in other exons or due to an admixture of a too high number of non-neoplastic cells in the cores selected for sequencing. Furthermore, using microdissection to exclude admixture of stroma cells, Griewe et al,26 had previously found p53 mutations in only 69% of p53-positive cases. It cannot be excluded that some p53-positive cases without detectable mutation in our study and in the previous work by Griewe may have mutations in other exons than the hot-spot exons 4–8. Other studies had also reported low frequencies of p53 positivity by immunohistochemistry. Visakorpi et al,27 found 6% p53-positive cases in a study of one large section per prostate cancer. Zellweger et al,6 detected 4% p53-positive prostate cancers in a tissue-microarray analysis of 181 patients. The majority of studies finding frequencies of p53 alterations between 2 and 20% describe association with unfavorable phenotype and/or poor prognosis as observed in our study.6, 27, 28
The higher likelihood of positive immunostainings (false and true) in larger sized tissue samples makes it difficult to use prognostic associations of molecular features identified in tissue-microarray studies for clinical routine.25 Extensive validation experiments and, potentially also, adjustments of protocols and criteria for ‘positivity’ may be required before routine use affecting clinical decision-making. It is important to note that in the case of prostate cancer there may be an exception from this rule. In this cancer, initial diagnosis is typically made on very small tissue samples, for which tissue microarrays may represent an ideal model for routine molecular-analysis simulation. The amount of tumor available for molecular analysis on needle core biopsies is approximately comparable to the situation on tissue microarrays. Therefore, it can be speculated that prognostic biomarkers identified on tissue microarrays may be transferable to needle core biopsies and thus be utilized for an improved preoperative risk assessment with potential importance on the therapeutic decision-making.
In summary, the results of this study show that p53 mutations characterize a small biologically aggressive subgroup with a high risk of progression after radical prostatectomy. As it is particularly important to find a biomarker for identifying low- and intermediate-grade cancers with an aggressive clinical course, it is interesting that we could demonstrate that p53 expression is an independent predictor of PSA recurrence in these cancers. Nevertheless, p53 alterations are very rare in this group of cancers. Further studies are needed to determine whether p53-positive prostate cancers should undergo adjusted treatment strategies, potentially including neoadjuvant and/or adjuvant therapy.
References
Cox LS . Multiple pathways control cell growth and transformation: overlapping and independent activities of p53 and p21Cip1/WAF1/Sdi1. J Pathol 1997;183:134–140.
Bosari S, Viale G . The clinical significance of p53 aberrations in human tumours. Virchows Arch 1995;427:229–241.
Erbersdobler A, Fritz H, Schnoger S, et al. Tumour grade, proliferation, apoptosis, microvessel density, p53, and bcl-2 in prostate cancers: differences between tumours located in the transition zone and in the peripheral zone. Eur Urol 2002;41:40–46.
Navone NM, Troncoso P, Pisters LL, et al. p53 protein accumulation and gene mutation in the progression of human prostate carcinoma. J Natl Cancer Inst 1993;85:1657–1669.
Quinn DI, Henshall SM, Head DR, et al. Prognostic significance of p53 nuclear accumulation in localized prostate cancer treated with radical prostatectomy. Cancer Res 2000;60:1585–1594.
Zellweger T, Ninck C, Bloch M, et al. Expression patterns of potential therapeutic targets in prostate cancer. Int J Cancer 2005;113:619–628.
Augustin H, Hammerer PG, Graefen M, et al. Characterisation of biomolecular profiles in primary high-grade prostate cancer treated by radical prostatectomy. J Cancer Res Clin Oncol 2003;129:662–668.
Grignon DJ, Caplan R, Sarkar FH, et al. p53 status and prognosis of locally advanced prostatic adenocarcinoma: a study based on RTOG 8610. J Natl Cancer Inst 1997;89:158–165.
Stapleton AM, Zbell P, Kattan MW, et al. Assessment of the biologic markers p53, Ki-67, and apoptotic index as predictive indicators of prostate carcinoma recurrence after surgery. Cancer 1998;82:168–175.
Scherr DS, Vaughan Jr ED, Wei J, et al. BCL-2 and p53 expression in clinically localized prostate cancer predicts response to external beam radiotherapy. J Urol 1999;162:12–16; discussion 16–17.
Borre M, Stausbol-Gron B, Overgaard J . p53 accumulation associated with bcl-2, the proliferation marker MIB-1 and survival in patients with prostate cancer subjected to watchful waiting. J Urol 2000;164:716–721.
Brooks JD, Bova GS, Ewing CM, et al. An uncertain role for p53 gene alterations in human prostate cancers. Cancer Res 1996;56:3814–3822.
Uzoaru I, Rubenstein M, Mirochnik Y, et al. An evaluation of the markers p53 and Ki-67 for their predictive value in prostate cancer. J Surg Oncol 1998;67:33–37.
Salem CE, Tomasic NA, Elmajian DA, et al. p53 protein and gene alterations in pathological stage C prostate carcinoma. J Urol 1997;158:510–514.
Claudio PP, Zamparelli A, Garcia FU, et al. Expression of cell-cycle-regulated proteins pRb2/p130, p107, p27(kip1), p53, mdm-2, and Ki-67 (MIB-1) in prostatic gland adenocarcinoma. Clin Cancer Res 2002;8:1808–1815.
Bauer JJ, Sesterhenn IA, Mostofi KF, et al. p53 nuclear protein expression is an independent prognostic marker in clinically localized prostate cancer patients undergoing radical prostatectomy. Clin Cancer Res 1995;1:1295–1300.
Bubendorf L, Kononen J, Koivisto P, et al. Survey of gene amplifications during prostate cancer progression by high-throughout fluorescence in situ hybridization on tissue microarrays. Cancer Res 1999;59:803–806.
Heidenreich A, Aus G, Bolla M, et al. EAU guidelines on prostate cancer. Eur Urol 2008;53:68–80.
Nesslinger NJ, Shi XB, deVere White RW . Androgen-independent growth of LNCaP prostate cancer cells is mediated by gain-of-function mutant p53. Cancer Res 2003;63:2228–2233.
Tepper CG, Gregg JP, Shi XB, et al. Profiling of gene expression changes caused by p53 gain-of-function mutant alleles in prostate cancer cells. Prostate 2005;65:375–389.
Han M, Partin AW, Pound CR, et al. Long-term biochemical disease-free and cancer-specific survival following anatomic radical retropubic prostatectomy. The 15-year Johns Hopkins experience. Urol Clin North Am 2001;28:555–565.
Rubin MA, Dunn R, Strawderman M, et al. Tissue microarray sampling strategy for prostate cancer biomarker analysis. Am J Surg Pathol 2002;26:312–319.
Sauter G, Simon R, Hillan K . Tissue microarrays in drug discovery. Nat Rev Drug Discov 2003;2:962–972.
Chen Z, Trotman LC, Shaffer D, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 2005;436:725–730.
Torhorst J, Bucher C, Kononen J, et al. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol 2001;159:2249–2256.
Griewe GL, Dean RC, Zhang W, et al. p53 Immunostaining guided laser capture microdissection (p53-LCM) defines the presence of p53 gene mutations in focal regions of primary prostate cancer positive for p53 protein. Prostate Cancer Prostatic Dis 2003;6:281–285.
Visakorpi T, Kallioniemi OP, Heikkinen A, et al. Small subgroup of aggressive, highly proliferative prostatic carcinomas defined by p53 accumulation. J Natl Cancer Inst 1992;84:883–887.
Henke RP, Kruger E, Ayhan N, et al. Immunohistochemical detection of p53 protein in human prostatic cancer. J Urol 1994;152:1297–1301.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Schlomm, T., Iwers, L., Kirstein, P. et al. Clinical significance of p53 alterations in surgically treated prostate cancers. Mod Pathol 21, 1371–1378 (2008). https://doi.org/10.1038/modpathol.2008.104
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2008.104
Keywords
This article is cited by
-
Focal p53 protein expression and lymphovascular invasion in primary prostate tumors predict metastatic progression
Scientific Reports (2022)
-
Biologically informed deep neural network for prostate cancer discovery
Nature (2021)
-
Subcellular Compartmentalization of Survivin is Associated with Biological Aggressiveness and Prognosis in Prostate Cancer
Scientific Reports (2020)
-
Upregulation of the heterogeneous nuclear ribonucleoprotein hnRNPA1 is an independent predictor of early biochemical recurrence in TMPRSS2:ERG fusion-negative prostate cancers
Virchows Archiv (2020)
-
Secreted Frizzled-Related Protein 4 (SFRP4) Is an Independent Prognostic Marker in Prostate Cancers Lacking TMPRSS2: ERG Fusions
Pathology & Oncology Research (2020)