Abstract
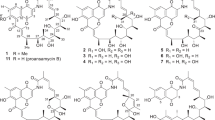
Seventeen new compounds, naphthacemycins A1–A11, B1–B4 and C1–C2, were isolated from a cultured broth of Streptomyces sp. KB-3346-5 during screening for circumventors of β-lactam resistance in methicillin-resistant Staphylococcus aureus. Their structures were elucidated by spectroscopic studies, including NMR and X-ray crystallographic analysis. The naphthacemycin A series has a new skeleton displaying a 7-phenylnaphthacene-5,6,11(12H)-trione. In contrast, the quinone moiety of the A series is changed to dehydroxyquinol in the B series and to a semiquinone-like structure in the C series.
Similar content being viewed by others
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of untreatable and potentially fatal hospital-associated infections. Community-acquired MRSA has also become a serious public health issue.1 MRSA is resistant to β-lactam antibiotics and usually resistant to most other classes of antibiotics. There are a few antibiotics used for MRSA, for example, vancomycin, teicoplanin, arbekacin, linezolid, daptomycin and tigecycline, but microorganism strains resistant to these compounds are increasingly being reported. In the course of screening for new anti-MRSA compounds, we found cyslabdan, which enhances the activity of imipenem (a carbapenem type β-lactam antibiotic) against MRSA.2, 3 Further screening for microbial metabolites that circumvent the β-lactam resistance of MRSA led us to find the naphthacemycins A1–A11 (1–11), B1–B4 (12–15) and C1–C2 (16–17) (Scheme 1), reported as KB-3346-5 substances in the patent by our group,4 from a cultured broth of Streptomyces sp. KB-3346-5.5 Here we describe the structure elucidation of the naphthacemycins, based on NMR study and X-ray crystallographic analysis.
Results and discussion
Physicochemical properties
Naphthacemycins (1–17) were purified from a cultured broth of Streptomyces sp. KB-3346-5, isolated from a soil sample collected in Okinawa Prefecture, Japan.5 They are red powders. Compounds 2, 4-6, 8, 11 and 14–17 have one chlorine atom, while 7 and 10 have two chlorine atoms. The IR spectra showed they all have carbonyl (1600–1720 cm−1) and hydroxyl (3350–3440 cm−1) groups. UV spectra were observed at 274–280, 302–306 and 362–408 nm in the naphthacemycin A series (1–11), at 246–249, 287–288, 352–354 and 414–417 nm in the naphthacemycin B series (12–15), and 248 and 410 nm in the naphthacemycin C series (16–17). Naphthacemycins are soluble in chloroform, ethyl acetate and methanol and insoluble in n-hexane and H2O.
Structure elucidation of the naphthacemycin A series
The molecular formula of naphthacemycin A9 (9) was established as C30H26O8 by HR-FAB-MS. 1H and 13C NMR spectra and HSQC analysis revealed the presence of 30 carbons, including six methyl, six sp2 methine, one sp3 quaternary and 17 sp2 quaternary carbons (Table 1). The long-range couplings of HMBC correlation revealed fragments I and II (Figure 1) as follows; the cross peaks from H-1 (δH 6.55) to C-2 (δC 163.9), C-3 (δC 101.6), C-4a (δC 110.0) and C-12a (δC 154.7), from H-3 (δH 6.25) to C-1 (δC 105.5), C-2, C-4 (δC 165.0) and C-4a, and from 4-OH (δH 12.86) to C-3, C-4 and C-4a indicated a 2,3,5-trisubstituted phenol (ring A). The cross peaks from H3-13 (δH 1.77) and H3-14 (δH 1.83) to C-11a (δC 155.4), C-12 (δC 39.2) and C-12a indicated a dimethyl moiety being connected to C-12a via C-12. This was confirmed by the coupling between H-1 and C-12, and thus the fragment I was established. An oxygen atom is suggested to be attached to C-2 by its chemical shift. The cross peaks from H-8 (δH 6.99) to C-6a (δC 125.8), C-7 (δC 140.1), C-9 (δC 162.6) and C-10 (δC 109.5), from H-10 (δH 7.52) to C-6a, C-8 (δC 124.6), C-9, C-10a (δC 135.8) and C-11 (δC 185.7), and from 9-OCH3 (δH 3.94) to C-9 indicated a 3,4,5-trisubstituted anisole (ring D). The cross peaks from H-17 (δH 6.39) to C-15 (δC 120.9), C-16 (δC 156.7), C-18 (δC 159.9) and C-19 (δC 106.9), from H-19 (δH 6.44) to C-15, C-17 (δC 96.4) and C-18, from H3-21 (δH 2.06) to C-15, C-19 and C-20 (δC 137.0), from 16-OCH3 (δH 3.65) to C-16, and from 18-OCH3 (δH 3.80) to C-18 indicated a 2-monosubstituted 3,5-dimethoxytoluene (ring E). The coupling between H-8 and C-15 suggested a connection of rings D and E at C-7 and C-15, respectively, and thus the fragment II was established. Among the remaining three carbons, C-5 (δC 185.0) had couplings with H-1 and H-3 and C-6 (δC 182.6) had a coupling with H-10. They are considered to be 4JCH W-couplings, and C-5 and C-6 are suggested to bond to C-4a and C-6a, respectively (Figure 2). The chemical shifts of C-6 (δC 182.6) and C-11 (δC 185.7) indicated that they form a quinone. If the remaining C-5a (δC 135.5) bonds to C-5 and C-11a to form ring B, and C-5a–C-6 and C-11–C-11a bondings form ring C, the naphthacenequinone structure is constructed. C-2 is suggested to be hydroxylated by the molecular formula. The carbon connections unrevealed by HMBC were clarified by INADEQUATE analysis (Table 1). Thus the structure of 9 was elucidated as shown in Scheme 1. It has a 7-phenylnaphthacene-5,6,11(12H)-trione skeleton. Most structurally related compounds, tetarimycin A6 and fasamycins7, were recently reported by Brady and co-workers as antibacterial agents. The structure was confirmed by X-ray crystallographic analysis as shown in Figure 3. The naphthacenequinone forms a planar skeleton.
The molecular formula of naphthacemycin A8 (8) was established as C30H25ClO8 by HR-FAB-MS, and this suggested that 8 was chlorinated analog of 9. 1H and 13C NMR spectra of 8 were similar to those of 9, except for ring D (Table 1). The HMBC correlations from H-8 (δH 6.93) to C-6a (δC 127.1), C-7 (δC 137.4), C-9 (δC 158.4) and C-10 (δC 120.4) and from 9-OCH3 (δH 3.95) to C-9 were observed, and C-10 was a quaternary carbon (Figure 4). Thus, the structure of 8 was elucidated as 10-chloro-9, which was confirmed by X-ray crystallographic analysis (Figure 5). In contrast to 9, the naphthacenequinone of 8 bent about 40° at two quinone carbonyl atoms of ring C.
The structures of the other naphthacemycin A series were elucidated by comparison with NMR data of 8 or 9 or other A series compounds (Supplementary Tables S1 and S2) and analyses of HMBC correlations (Supplementary Figure S1).
Structure elucidation of the naphthacemycin B series
The molecular formula of naphthacemycin B1 (12) was established as C27H22O7 by HR-FAB-MS, which indicated 12 was 28 mass units (CO) less than 1. 1H and 13C NMR spectra and HSQC analysis revealed the presence of 27 carbons, including three methyl, seven sp2 methine, one sp3 quaternary and sixteen sp2 quaternary carbons (Table 1). The structure was elucidated by long-range couplings of HMBC correlation (Figure 4). The cross peaks from H-1 (δH 6.65) to C-2 (δC 166.7), C-3 (δC 102.1) and C-4a (δC 108.6), from H-3 (δH 6.20) to C-1 (δC 107.1), C-2, C-4 (δC 166.6) and C-4a, and from 4-OH (δH 12.87) to C-3, C-4 and C-4a indicated a 2,3,5-trisubstituted phenol (ring A). The cross peaks from H3-13 (δH 1.91) and H3-14 (δH 1.70) to C-11a (δC 146.5), C-12 (δC 39.7) and C-12a (δC 155.5) indicated a dimethyl moiety connected to C-12a via C-12. This was confirmed by the coupling between H-1 and C-12. The cross peaks from H-8 (δH 6.72) to C-6a (δC 118.2), C-9 (δC 166.9) and C-10 (δC 110.1) and from H-10 (δH 7.05) to C-6a, C-8 (δC 122.5), C-9, C-10a (δC 142.9) and C-11 (δC 116.1) indicated a 3,4,5-trisubstituted phenol (ring D). The cross peaks from H-11 (δH 7.36) to C-5a (δC 107.6), C-6a and C-12 and from 6-OH (δH 14.53) to C-5a, C-6 (δC 160.1) and C-6a indicated a 2,3,5,6-tetrasubstituted phenol (ring C), which condensed with ring D and connected to ring A via C-12. Comparing NMR data and structures of 12 with 1, it is appropriate to form 4,4-dimethylcyclohexadienone (ring B) by C-4a, 5, 5a, 11a, 12 and 12a, though C-5 had no coupling with neighboring protons. The resemblance of the NMR data also suggested that a hydroxyl residue is attached to C-2 of 12 as in 1. The cross peaks from H-17 (δH 6.23) to C-15 (δC 124.5), C-16 (δC 155.4), C-18 (δC 157.5) and C-19 (δC 108.8), from H-19 (δH 6.26) to C-15, C-17 (δC 100.8) and C-18, and from H3-21 (δH 1.88) to C-15, C-19 and C-20 (δC 138.3) indicated 2-monosubstituted 3,5-dihydroxytoluene (ring E). The coupling between H-8 and C-15 indicated that ring E connects to ring D at C-7. Thus, the structure of 12 was elucidated as 6,11-didehydro-O16-demethyl-6,11-dideoxo-11-hydroxy-1.
Structure elucidation of the naphthacemycin C series
The molecular formula of naphthacemycin C2 (17) was established as C33H31ClO9 by HR-FAB-MS, which indicated 17 was 58 mass units (C3H6O) more than 11. 1H and 13C NMR spectra and HSQC analysis revealed the presence of 33 carbons, including seven methyl, one sp3 methylene, five sp2 methine, two sp3 quaternary and eighteen sp2 quaternary carbons (Table 1). The structure was elucidated by long-range couplings of HMBC correlation (Figure 6). The cross peaks from H-1 (δH 6.46) to C-2 (δC 162.6), C-3 (δC 102.1) and C-4a (δC 108.6), from H-3 (δH 6.26) to C-1 (δC 105.4), C-2, C-4 (δC 165.4) and C-4a, and from 4-OH (δH 12.52) to C-3, C-4 and C-4a indicated a 2,3,5-trisubstituted phenol (ring A). The cross peaks from H3-13 (δH 1.59) and H3-14 (δH 1.01) to C-11a (δC 53.2), C-12 (δC 43.3) and C-12a (δC 152.9) indicated a dimethyl moiety was connected to C-12a via C-12. This was confirmed by the coupling between H-1 and C-12. The cross peaks from H-8 (δH 6.98) to C-6a (δC123.8), C-9 (δC 161.9) and C-10 (δC 110.1), H-10 (δH 7.73) to C-6a, C-8 (δC 124.6), C-9, C-10a (δC 137.3) and C-11 (δC 197.8), and from 9-OCH3 (δH 3.94) to C-9 indicated a 3,4,5-trisubstituted anisole (ring D). The cross peaks from 6-OH (δH 14.74) to C-5a (δC 107.6), C-6 (δC 168.8) and C-6a and from H2-22 (δH 2.89, 3.52) to C-5a, C-11 and C-11a indicated a 2,3,5,6-tetrasubstituted 4-hydroxy-2,4-cyclohexadienone (ring C), condensed with ring D and connected to ring A via C-12. The cross peaks from H3-24 (δH 1.89) to C-22 (δC 52.4) and C-23 (δC 205.4) and from H2-22 to C-23 indicated that a 2-oxopropyl residue was attached to C-11a. Comparing NMR data and structures of 17 with 11, it is appropriate to form 4,4-dimethyl-2-cyclohexenone (ring B) by C-4a, 5, 5a, 11a, 12 and 12a, though C-5 had no coupling with neighboring protons. The chemical shifts of ring E of 17 is quite similar to 11, which suggests ring E of 17 is 2-monosubstituted 4-chloro-3,5-dimethoxytoluene. This was confirmed by the cross peaks from H-19 (δH 6.61) to C-15 (δC 130.2), C-17 (δC 113.5) and C-18 (δC 154.7), from H3-21 (δH 1.96) to C-15, C-19 (δC 108.6) and C-20 (δC 134.9), from 16-OCH3 (δH 3.63) to C-16 (δC 153.1), and from 18-OCH3 (δH 3.94) to C-18. The coupling between H-8 and C-15 indicated that ring E connects to ring D at C-7. Thus, C-11a of 11 was substituted with a 2-oxopropyl group and the C-6 ketone was reduced in 17.
The molecular formula of naphthacemycin C1 (16) was established as C32H29ClO9 by HR-FAB-MS, which indicated 16 was 14 mass units (CH2) less than 17. 1H and 13C NMR spectra of 16 were quite similar to those of 17, except for rings D and E (Supplementary Table S3). The H-10 signal of 17 disappeared in 16 and the cross peaks of HMBC from H-8 (δH 7.01) to C-6a (δC 126.7), C-9 (δC 158.6) and C-10 (δC 120.8) and from 9-OCH3 (δH 4.03) to C-9 were observed, which indicated that a chlorine was attached to the C-10 of ring D (Figure 6). The 18-OCH3 signal of 17 was also absent in 16 and an H-17 (δH 6.37) signal appeared in 17. The cross peaks from H-17 (δH 6.37) to C-15 (δC 123.7), C-16 (δC 158.1), C-18 (δC 158.4) and C-19 (δC 109.4), from H-19 (δH 6.39) to C-15, C-17 (δC 97.4) and C-18, from H3-21 (δH 2.01) to C-15, C-19 and C-20 (δC 137.0), and from 16-OCH3 (δH 3.64) to C-16 indicated a ring E structure of 2-monosubstituted 3,5-dimethoxytoluene. Thus 16 was 10-chloro-17-dechloro-O18-demethyl-17. We have not elucidated the configurations of 16 and 17 yet.
Circumvention of β-lactam resistance in MRSA
The circumvention of β-lactam resistance in MRSA was measured by enhancement of imipenem activity on MRSA using the paper disk method. Naphthacemycins alone showed no antibacterial activity against a clinically isolated MRSA strain K24 at 0.01–1 μg per disk. When the agar plates contained 10 μg ml−1 of imipenem, which also did not affect the growth of MRSA, 0.01–1 μg per disks of naphthacemycins showed inhibition zones (Table 2). Compounds 3 and 5–10 inhibited the growth of MRSA at 0.01 μg per disk in the presence of imipenem. Among them, 6 and 10 showed the largest inhibition zones (11 mm) at 0.01 μg per disk. The detailed activity of naphthacemycins against MRSA will be reported in an accompanying paper.5
Conclusion
In conclusion, 17 new compounds designated naphthacemycins were isolated from the culture broth of Streptomyces sp. KB-3346-5. They are circumventors of β-lactam resistance and enhanced imipenem activity against β-lactam resistant MRSA. Naphthacemycins are 1-phenylnaphthacene antibiotics produced by Streptomyces sp. Many naphthacene type compounds have been isolated from actinomycetes, but most of them have partially unsaturated rings, such as tetracyclines and anthracyclines. Some highly unsaturated naphthacene compounds have been reported to be produced by actinomycetes, such as tetracenomycin D, galtamycin and tetracenoquinocin.8, 9, 10 A biosynthetic intermediate of tetracycline, pretetramid and an aglycone of anthracycline, η-pyrromycinone, are classified in the latter.11, 12 Naphthacemycins A and B series also belong to the latter group, but they are the first compounds having a 1-phenylnaphthacene skeleton isolated from a natural origin.
Methods
General experimental procedure
1H NMR (300 MHz) and 13C NMR (75 MHz) spectra were recorded on a Varian XL-300 spectrometer. Chemical shifts are shown in δ values (p.p.m.) relative to the solvents (acetone-d6 at 2.05 p.p.m. for 1H NMR and at 29.8 p.p.m. for 13C NMR; CDCl3 at 7.26 p.p.m. for 1H NMR and at 77.0 p.p.m. for 13C NMR; DMSO-d6 at 2.50 p.p.m. for 1H NMR and at 39.5 p.p.m. for 13C NMR). INADEQUATE experiment of 9 (170 mg) was carried out for 100 h by 125 MHz NMR using methanol-d4 as a solvent. Mass spectrometry was conducted on a JEOL JMS-AX505 HA spectrometer. The UV and IR spectra were measured with a Hitachi U-2810 spectrophotometer and a Horiba FT-710 Fourier transform infrared spectrometer, respectively. Optical rotations were recorded on a JASCO model DIP-1000 polarimeter.
Assay of antibacterial activity
Measurement of inhibition zone of naphthacemycins, with or without imipenem, to evaluate circumvention activity of β-lactam resistance were carried out by paper disk method, as reported previously.3
Data availability
The X-ray crystallographic data have been deposited at the Cambridge Crystallographic Data Centre (CCDC) under deposition numbers CCDC 1536220 (8) and CCDC 1536223 (9).

Structures of naphthacemycins A1–A11 (1–11), B1–B4 (12–15) and C1–C2 (16–17).
References
Diep, B. A. & Otto, M. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 16, 361–369 (2008).
Fukumoto, A. et al. Cyslabdan, a new potentiator of imipenem activity against methicillin-resistant Staphylococcus aureus, produced by Streptomyces sp. K04-0144. I. Taxonomy, fermentation, isolation and structural elucidation. J. Antibiot. 61, 1–6 (2008).
Fukumoto, A. et al. Cyslabdan, a new potentiator of imipenem activity against methicillin-resistant Staphylococcus aureus, produced by Streptomyces sp. K04-0144. II. Biological activities. J. Antibiot. 61, 7–10 (2008).
Ōmura, S. et al(Kitasato Institute, Japan; Kyowa Hakko Kirin Co., Ltd., Japan) KB-3346-5 substances, their fermentative manufacture, and antibacterial agents containing them. Jpn Kokai Tokkyo Koho JP2009046404A (2009).
Fukumoto, A. et al. Naphthacemycins, novel circumventors of β-lactam resistance in MRSA, produced by Streptomyces sp. KB-3346-5. I. The taxonomy of the producing strain, and the fermentation, isolation and antibacterial activities. J. Antibiot. (doi:10.1038/ja.2017.28).
Feng, Z., Kallifidas, D. & Brady, S. F. Functional analysis of environmental DNA-derived type II polyketide synthases reveals structurally diverse secondary metabolites. Proc. Natl Acad. USA 108, 12629–12634 (2011).
Feng, Z., Chakraborty, D., Dewell, S. B., Reddy, B. V. B. & Brady, S. F. Environmental DNA-encoded antibiotics fasamycins A and B inhibit FabF in type II fatty acid biosynthesis. J. Am. Chem. Soc. 134, 2981–2987 (2012).
Yue, S., Motamedi, H., Wendt-Pienkowski, E. & Hutchinson, C. R. Antracycline metabolites of tetracenomycin C-ninproducing Streptomyces glaucescens mutants. J. Bacteriol. 167, 581–586 (1986).
Egorov, L. V., Tetent’eva, T. G., Rudneva, N. A., Egorenko, G. G. & Ivantiskaia, L. P. Experimental study of the antitumor anthracycline antibiotic aclarubicin (aclacinomycin A). Antibiot. Med. Biotekhnol. 30, 918–927 (1985).
Motohashi, K., Takagi, M. & Shin-ya, K. Tetracenoquinocin and 5-iminoaranciamycin from a sponge-derived Streptomyces sp. Sp080513GE-26. J. Nat. Prod. 73, 755–758 (2010).
Zhang, W., Watanabe, K., Wang, C. C. & Tang, Y. Investigation of early tailoring reactions in the oxytetracycline biosynthetic pathway. J. Biol. Chem. 282, 25717–25725 (2007).
Brockmann, H., Pla, L. C. & Lenk, W. ζ-pyrromycinone. Angew. Chem. 69, 477 (1957).
Acknowledgements
This work was supported in part by a grant of the twenty-first century COE Program, Ministry of Education, Culture, Sports, Science and Technology, Japan. We are grateful to Ms Akiko Nakagawa and Ms Noriko Sato of the School of Pharmacy, Kitasato University, for measurements of mass and NMR spectra. We thank Varian Technologies Japan Ltd for the measurement of INADEQUATE.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Fukumoto, A., Kim, YP., Iwatsuki, M. et al. Naphthacemycins, novel circumventors of β-lactam resistance in MRSA, produced by Streptomyces sp. KB-3346-5. II. Structure elucidation. J Antibiot 70, 568–573 (2017). https://doi.org/10.1038/ja.2017.29
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2017.29
This article is cited by
-
Streptovertimycins A–H, new fasamycin-type antibiotics produced by a soil-derived Streptomyces morookaense strain
The Journal of Antibiotics (2020)
-
Naphthacemycins, novel circumventors of β-lactam resistance in MRSA, produced by Streptomyces sp. KB-3346-5. I. The taxonomy of the producing strain, and the fermentation, isolation and antibacterial activities
The Journal of Antibiotics (2017)