Abstract
Hypertensive disorders of pregnancy (HDP) is a common disease and is believed to be a multifactorial genetic disease. Stromal interaction molecule 1 (STIM1) was previously reported to regulate the concentration of Ca2+ and vascular contraction. The aim of the present study was to assess the association between HDP and single-nucleotide polymorphisms (SNPs) or haplotypes in the human STIM1 gene via case–control studies. On the basis of a database on the National Center of Biotechnology Information website, we selected five SNPs in the human STIM1 gene and performed an association study with 139 HDP patients and 162 age-matched non-HDP subjects. There were significant differences between the HDP and control groups in the genotypes (P=0.041) and recessive models (P=0.045) for rs7945554, and between the gestational hypertension and control groups in the dominant models (P=0.015) and alleles (P=0.043) for rs10458894. The haplotypes of A-T-G-G, A-C-A-G, A-T-A-G, G-T-G-C, A-T-G-C and G-C-A-C (rs7945554–rs10458894–rs7929653–rs2923956) were significantly different from those of the control group. In the logistic regression analysis, the AA genotype of rs7945554 was significantly more predominant in the HDP group than in the control group. We found HDP-sensitive SNPs and haplotypes, and the STIM1 gene was identified as a possible susceptibility gene for HDP. By providing guidance to patients with genetic factors for HDP, we may be able to help them avoid environmental factors that could increase the risk of HDP before or during pregnancy and thus prevent or delay the onset of the disease.
Similar content being viewed by others
Introduction
Hypertensive disorders of pregnancy (HDP) is a common disease that occurs in ~5–7% of pregnant women in Japan. Many causes of HDP have been reported, and it is believed to be a multifactorial genetic disease. Disorders of endothelial cells, thrombotic tendencies, dysfunctions of the placenta and oxygen deficiencies are the main mechanisms behind HDP.
In humans, Ca2+ has important roles in immunity for second messengers, cell proliferation, secretion, blood coagulation, muscle contraction and neurotransmission.1 Ca2+ exists outside of cells or is stored in organelles, such as the endoplasmic reticulum (ER). Extracellular stimulation can cause a temporary release of Ca2+ from the ER and sustained Ca2+ entry into cells via calcium channels; this is called store-operated calcium entry (SOCE), also known as capacitative calcium entry or calcium-release-activated calcium entry (CRAC).2 Two proteins have important roles in SOCE: CRAC modulator 13, 4 (Orai1, also called CRACM1) and stromal interaction molecule 15, 6 (STIM1).
Orai1 is a four-pass transmembrane calcium channel protein found in the cell membrane, with its N terminus and C terminus in the intracytoplasmic space. STIM1 is a single-pass transmembrane protein that is located in the ER membrane and its N terminus is in the ER lumen.
When the concentration of Ca2+ decreases in the ER, the EF hand motif of STIM1 senses this concentration change, and Orai1 and STIM1 form clusters. These clusters activate SOCE, and Ca2+ flows in from the outside of the cell.7 This system has been confirmed in immune cells, muscular cells, nerve cells, internal secretion cells and cancer cells. STIM proteins have two isoforms (STIM1 and STIM2), and STIM2 is reported to be related to SOCE. However, most aspects of the role of STIM2 in SOCE-related diseases remain unknown.
Heredity is known to be one of the risk factors for HDP, and associations between HDP and various genes have been reported in many races.8, 9 STIM1 and Orai1 gene variations have been reported to inhibit SOCE and to cause severe combined immunodeficiency.3 SOCE may contribute greatly to the immune system. Mice lacking STIM1 or STIM2 were reported to have particularly low levels of regulatory T cells among the T cells.10 In HDP patients, the number of T cells is decreased in the peripheral blood, causing the collapse of immune tolerance to fetus cells and the development of HDP.11 However, to date the associations between STIM1 genes and HDP have not been reported.
Levels of STIM1 and Orai1 have been reported to be increased in the vascular smooth muscle cells of hypertension model rats.12 It is possible that SOCE contributes to the pathology of hypertension in numerous ways.
The aim of the present study was to assess the association between HDP and single-nucleotide polymorphisms (SNPs) or haplotypes in the human STIM1 gene via case–control studies.
Methods
Subjects
We collected samples from 139 Japanese women with a history of HDP, and 162 Japanese women with no history of HDP as controls, from 2006 to 2015. All participants had attended the Department of Obstetrics and Gynecology, Nihon University School of Medicine Itabashi Hospital, Tokyo, Japan. Women with multiple pregnancies or a history of essential hypertension before pregnancy were excluded from both groups. Samples were collected from participants of whom 1.19% were in their sixties, 0.79% were in their fifties, 6.72% were in their forties, 54.2% were in their thirties, 40.3% were in their twenties and 1.19% were in their teens. We collected the information from participants in their sixties and fifties from interviews and from maternal and child health handbooks, which have been published since the 1940s in Japan. As age-matching is the most important condition of a case–control study, the age at delivery for both groups was matched in the present study. The HDP group was subdivided into two groups: those with preeclampsia (PE) and those with gestational hypertension (GH).
HDP was defined as a systolic blood pressure ⩾140 mm Hg or a diastolic blood pressure ⩾90 mm Hg that developed after 20 weeks of gestation and improved by 12 weeks postpartum. PE was defined as hypertension with proteinuria that developed after 20 weeks of gestation and improved by 12 weeks postpartum. GH was defined as hypertension without proteinuria that occurred after 20 weeks of gestation and developed by 12 weeks postpartum. Proteinuria was defined as protein excretion ⩾300 mg in a 24-h urine specimen collection.13
We selected subjects in accordance with the criteria of GH and PE, and the presence of renal diseases or diabetes mellitus was not included. In the control group, 1.2% had renal diseases and 0.6% had diabetes mellitus, excluding gestational diabetes mellitus. In the HDP group 2.8% had renal diseases and none had diabetes mellitus. As there were no significant differences in the presence of these diseases between the two groups, we consider that the choice of participants was reasonable.
Informed consent was obtained from each participant. This study was approved by the Human Studies Committee of Nihon University, School of Medicine.
Genotyping
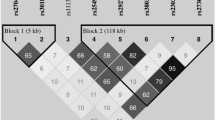
The STIM1 gene is located on chromosome 11p15.5, and it contains 13 exons and 238,684 bases (Figure 1).
We chose five SNPs (rs7945554, rs10458894, rs7929653, rs2923956 and rs10835596) in the STIM1 gene that have a minor allele frequency (MAF) of over 5% in the Japanese population. All SNPs were selected from the HapMap database (http://hapmap.ncbi.nlm.nih.gov/). The MAF and the nucleotide sequences around each SNP were as follows: rs7945554, MAF (A)=0.25, 5′-GTCAGAAAAGGAAATATAGGTCATG[A/G]TGGCTGGGGAGTTTCATGGATGGAT-3′; rs10458894, MAF (T)=0.34, 5′-AGTGTCTAAGGACAAAATAGAAATT[C/T]GGTAAATGTTTGTTGAATTGTAGAC-3′;rs7929653, MAF (A)=0.31, 5′-CGATATTTAAGATGGGTGTGACATG[A/G]CCAGAGTGTTTTTTTTAGGAAGACC-3′; rs2923956, MAF (C)=0.50, 5′-GCCCACCTCAGCCTCCCAAAATCCT[C/G]GGATCGTAAACATGAGCCAACGCA-3′; and rs10835596, MAF (T)=0.32, 5′-GGTCCAGCCCAATTTTCGGCTGTCA[C/T]CTAAAATCATCAATGTCTCCCAGGA-3′.
Genomic DNA was obtained from peripheral blood mononuclear cells using phenol–chloroform extraction and ethanol precipitation. The concentration of DNA was adjusted to 1 ng μl−1 using a spectrophotometer.14 We identified the genotypes using TaqMan PCR with Assays-on-Demand kits (Applied Biosystems, Foster City, CA, USA).
For each reaction, we mixed 2 μl of DNA, 2.5 μl of TaqMan Genotyping Master Mix (Applied Biosystems), 0.16 μl of Tris-EDTA buffer 1 × (Promega, Fitchburg, WI, USA), 2.25 μl of distilled water as primer and 0.0092 μl of TaqMan SNP Genotyping Assays (Applied Biosystems) as probe. Each PCR reaction was performed in a well of a 96-well plate that was placed in a 2720 Thermal Cycler (Applied Biosystems). Thermal cycling was set to 95 °C for 10 min, and then 50 cycles of 92 °C for 15 s and 60 °C for 1 min, and, finally, reactions were held at 4 °C.14
We used an ABI PRISM 7700 Sequence Detector (Applied Biosystems) as a fluorescence detector for reading the end point of the TaqMan PCR, and the obtained data were analyzed using Detector v. 1.7 alias (Applied Biosystems).
Statistical analysis
We performed association studies (case–control studies) with 139 HDP patients and 162 age-matched non-HDP subjects using individual SNPs and haplotypes constructed with these SNPs. Continuous variables are shown as the mean±s.d. Differences in continuous variables between the HDP and control participants were analyzed with the Mann–Whitney U-test. Differences in categorical variables were analyzed with Fisher’s exact test. Statistical analysis was performed using GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA).
We carried out a linkage disequilibrium analysis and a haplotype-based case–control study using the SNPAlyze software program, version 3.2.3 (Dynacom, Yokohama, Japan), with four SNPs that conformed to Hardy–Weinberg equilibrium. Hardy–Weinberg equilibrium was calculated with the χ2-test, and the equilibrium was judged to be accurate with P-values >0.05 (rs12313273, P=0.92; rs6486795, P=0.62; rs7945554, P=0.27; rs10458894, P=0.29; rs7929653, P=0.10; and rs2923956, P=0.08). rs10835596 was excluded from the haplotype-based case–control study because it was not in Hardy–Weinberg equilibrium. All SNPs were confirmed to be on the same haplotype block by calculating the |D’ |values, and they were included in the haplotype-based case–control study, because all r2 values were >0.5 (Table 1). In the haplotype-based case–control study, haplotypes with frequencies <0.03 were excluded from the study. The distributions of haplotype frequencies were calculated using the χ2-test. Probability levels of P<0.05 were considered to indicate differences that are statistically significant.
Statistical analyses were performed using SPSS software for Windows, version 12 (SPSS, Chicago, IL, USA).
Results
The characteristics of the study participants are shown in Table 2. The proportion of those with a family history of hypertension, their systolic blood pressure, diastolic blood pressure and body mass index (BMI) before pregnancy, and their BMI at delivery, were higher than those in the control group. The gestational age at delivery, birth weight of neonates and Apgar score at 5 min were lower in the HDP and PE groups than those in the control group. The systolic blood pressure, diastolic blood pressure and BMI before pregnancy, and BMI at delivery, were higher, and the gestational age at delivery was lower in the GH group than in the control group.
The genotypes, dominant models, recessive models and alleles are indicated in Table 3. There were significant differences between the HDP and control groups in the genotypes (P=0.041) and recessive models (P=0.045) for rs7945554, and between the GH and control groups in the dominant models (P=0.015) and alleles (P=0.043) for rs10458894.
The haplotype-based control study results are shown in Table 4. The haplotypes of A-T-G-G, A-C-A-G, A-T-A-G, G-T-G-C, A-T-G-C and G-C-A-C (rs7945554–rs10458894–rs7929653–rs2923956) were significantly different from those of the control group.
For the logistic regression analysis, we chose age and a family history of hypertension as confounding factors. Confounding factors such as blood pressure, and BMI associated with criteria for HDP, were not estimated in this analysis. In the logistic analysis, we excluded factors included in the criteria of the disease for the confounding factors because these showed significant differences as a necessary consequence. The AA genotype of rs7945554 was significantly more predominant in the HDP group than in the control group (Table 5).
Discussion
A family history of hypertension, history of HDP, primipara status, obesity, elderly pregnancy, stress, diabetes, dental cavities, urinary tract infections and thyroid disease are the known risk factors of HDP. In this study, there were significant differences in the number of individuals with a family history of hypertension and in BMI, but the ages at delivery and the proportions of primigravidae did not differ significantly between the HDP and control groups. We saw differences in the gestational age at delivery, birth weight of neonates and Apgar score at 5 min between the HDP and control groups, because we chose to terminate pregnancy as a treatment for HDP, which leads to premature birth.
Although an association between STIM1 and hypertension has been reported using vascular smooth muscle cells of model rats, the present investigation is the first reported association study between HDP and the STIM1 gene. Association studies have high statistical power, and it is easy to collect samples; however, false-positive results are not uncommon. Genome-wide association studies (GWAS) are currently attracting much attention, and the inhibin βB gene15 and the PSG11 gene16 have been identified as genes associated with HDP in GWAS. We would have performed a GWAS had funding permitted; however, even if we had detected a susceptibility gene using GWAS, we would have subsequently have had to perform an association study comparable to the present study.
We found significant differences in the genotypes and alleles of two SNPs between the HDP and control participants. In rs7945554, we found differences between the HDP and control groups, but not between the PE or GH and control groups. rs7945554 may be associated with the development of HDP. In contrast, in rs10458894, we found differences between the GH and control groups, but not between the PE or HPD and control groups. Several theories speculate that PE and GH are involved in different pathologies and show different gene expression patterns. Proteins related to the pathology of HDP may cause differences, and some studies have reported genetic mutations in PE groups but not in GH groups.17, 18 In this study, the number of participants in the GH group was small, and this may account for the difference seen between the GH and control groups. The frequency of GH was consistently low, ranging from 0.4 to 0.6%. On the other hand, the frequency of PE was 2–3%. The period of sampling was inconsistent among participants. However, as the aim of this study was to investigate inheritable factors of HDP, we consider that the sampling period did not affect the results.
SNPs located in introns do not directly affect the expression of a protein. SNPs with individual differences were chosen as genetic markers; therefore, SNPs that did not show a significant effect in the association study could not be definitively ruled out as having an association with the disease.
Even if significant associations between a disease and a polymorphism are found, the polymorphism cannot definitively be said to confer disease susceptibility. Other polymorphisms in the same gene or neighboring genes may primarily determine susceptibility, and the observed associations may be secondary phenomena caused by linkage disequilibrium. We need to determine the region with the strongest association with a disease by conducting an association study of several of the nearby polymorphisms. For such a study, haplotype analysis would be useful. Gene recombination can occur at the time of hereditary transmission, and nearby SNPs are inherited as a group. This alignment of genes grouped on one chromosome is called a haplotype. The analysis of haplotypes may allow the determination of associations that were not detectable from the analysis of SNPs alone. The haplotype-based case–control study showed significant differences, indicating that the haplotypes were related to HDP. A-T-G-G, A-T-A-G, A-T-G-C and G-C-A-C could be disease-susceptibility haplotypes, whereas A-C-A-G and G-T-G-C could be disease-resistance haplotypes.
Associations have been reported between gene polymorphisms of the STIM1 gene and ankylosing spondylitis.19 We did not choose the same SNPs as used by those researchers because the MAFs of the SNPs are low in the Japanese population. However, we plan to use these SNPs in a future study.
In this study, we examined only maternal genetic backgrounds, but paternal genetic backgrounds are also known to be associated with the development of HDP. Having a father whose mother had HDP, or a father that fathered a child with another woman with HDP before fathering the participant in this study, is reported to increase the risk of HDP.20, 21 Cnattinguis et al.22 reported that the percentages of PE risk factors were attributable as follows: 35% to maternal genetic factors, 20% to fetal genetic factors, 13% to couple factors and 32% to unknown factors. Unfortunately, we did not collect paternal blood samples in this study. We hope to clarify the genetic factors of HDP by conducting studies of both maternal and paternal genes in the future.
HDP is a multifactorial genetic disease, and no study definitively identifying a single gene as an HDP risk factor has been published. In conclusion, we found HDP-sensitive SNPs and haplotypes, and identified the STIM1 gene as a possible susceptibility gene for HDP. By providing guidance to patients with genetic factors for HDP, we may be able to help them avoid environmental factors that could increase the risk of HDP before or during pregnancy and thus prevent or delay the onset of the disease.
References
Berridge MJ, Lipp P, Bootman MD . The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol 2000; 1: 11–21.
Putney JW Jr. . A model for receptor-regulated calcium entry. Cell Calcium 1986; 7: 1–12.
Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A . A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 2006; 441: 179–185.
Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, Kinet J . CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 2006; 312: 1220–1223.
Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE Jr., Meyer T . STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol 2005; CB 15: 1235–1241.
Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, Velicelebi G, Stauderman KA, Stauderman STIM1 . an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol 2005; 169: 435–445.
Lewis RS . The molecular choreography of a store-operated calcium channel. Nature 2007; 446: 284–287.
Luizon MR, Belo VA, Palei AC, Amaral LM, Lacchini R, Sandrim VC, Duarte G, Cavalli RC, Tanus-Santos JE . Effects of NAMPT polymorphisms and haplotypes on circulating visfatin/NAMPT levels in hypertensive disorders of pregnancy. Hypertens Res 2015; 38: 361–366.
Zhan Y, Liu M, You Y, Zhang Y, Wang J, Wang X, Liu S, Liu X . Genetic variations in the vitamin-D receptor (VDR) gene in preeclampsia patients in the Chinese Han population. Hypertens Res 2015; 38: 513–517.
Ohara N, Yamasaki M, Morikawa H, Ueda Y, Mochizuki M . Dynamics of calcium metabolism and calcium-regulating hormones in pregnancy-induced hypertension. Nihon Naibunpi Gakkai Zasshi 1986; 62: 882–896.
Sasaki Y, Darmochwal-Kolarz D, Suzuki D, Sakai M, Ito M, Shima T, Shiozaki A, Rolinski J, Saito S . Proportion of peripheral blood and decidual CD4(+) CD25(bright) regulatory T cells in pre-eclampsia. Clin Exp Immunol 2007; 149: 139–145.
Giachini FR, Chiao CW, Carneiro FS, Lima VV, Carneiro ZN, Dorrance AM, Tostes RC, Webb RC . Increased activation of stromal interaction molecule-1/Orai-1 in aorta from hypertensive rats: a novel insight into vascular dysfunction. Hypertension 2009; 53: 409–416.
No authors listed. Report of the National High Blood Pressure Education Program Working Group on high blood pressure in pregnancy. Am J Obstet Gynecol 2000; 183: S1–S22.
Tamura M, Nakayama T, Sato I, Sato N, Izawa N, Hishiki M, Mizutani Y, Furuya K, Yamamoto T . Haplotype-based case-control study of estrogen receptor alpha (ESR1) gene and pregnancy-induced hypertension. Hypertens Res 2008; 31: 221–228.
Johnson MP, Brennecke SP, East CE, Goring HH, Kent JW Jr., Dyer TD, Said JM, Roten LT, Iversen AC, Abraham LJ, Heinonen S, Kajantie E, Kere J, Kivinen K, Pouta A, Laivuori H, Austgulen R, Blangero J, Moses EK . Genome-wide association scan identifies a risk locus for preeclampsia on 2q14, near the inhibin, beta B gene. PLoS ONE 2012; 7: e33666.
Zhao L, Triche EW, Walsh KM, Bracken MB, Saftlas AF, Hoh J, Dewan AT . Genome-wide association study identifies a maternal copy-number deletion in PSG11 enriched among preeclampsia patients. BMC Pregnancy Childbirth 2012; 12: 61.
Mando C, Antonazzo P, Tabano S, Zanutto S, Pileri P, Somigliana E, Colleoni F, Martinelli A, Zolin A, Benedetto C, Marozio L, Neri I, Facchinetti F, Miozzo M, Cetin I . Angiotensin-converting enzyme and adducin-1 polymorphisms in women with preeclampsia and gestational hypertension. Reprod Sci 2009; 16: 819–826.
Sandrim VC, Palei AC, Cavalli RC, Araujo FM, Ramos ES, Duarte G, Tanus-Santos JE . Vascular endothelial growth factor genotypes and haplotypes are associated with pre-eclampsia but not with gestational hypertension. Mol Hum Reprod 2009; 15: 115–120.
Wei JC, Hung KS, Hsu YW, Wong RH, Huang CH, Jan MS, Wu SJ, Juan YS, Chang WC . Genetic polymorphisms of stromal interaction molecule 1 associated with the erythrocyte sedimentation rate and C-reactive protein in HLA-B27 positive ankylosing spondylitis patients. PLoS ONE 2012; 7: e49698.
Esplin MS, Fausett MB, Fraser A, Kerber R, Mineau G, Carrillo J, Varner MW . Paternal and maternal components of the predisposition to preeclampsia. N Engl J Med 2001; 344: 867–872.
Lie RT, Rasmussen S, Brunborg H, Gjessing HK, Lie-Nielsen E, Irgens LM . Fetal and maternal contributions to risk of pre-eclampsia: population based study. Br Med J 1998; 316: 1343–1347.
Cnattingius S, Reilly M, Pawitan Y, Lichtenstein P . Maternal and fetal genetic factors account for most of familial aggregation of preeclampsia: a population-based Swedish cohort study. Am J Med Genet A 2004; 130a: 365–371.
Acknowledgements
We wish to thank Ms K Sugama for her excellent technical assistance. This work was supported by a grant from the Health Sciences Research Institute, Yokohama, Japan, to the Division of Companion Diagnostics, Department of Pathology and Microbiology, Nihon University School of Medicine, Tokyo, Japan. This work was supported by JSPS KAKENHI Grant Number JP25460703.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
TN received 1 000 000 yen or more (in 1 year) for holding an advisory position. The remaining authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Shinya, K., Nakayama, T., Nakayama, T. et al. A case–control study between the STIM1 gene and hypertensive disorders of pregnancy. Hypertens Res 41, 39–44 (2018). https://doi.org/10.1038/hr.2017.84
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hr.2017.84
Keywords
This article is cited by
-
A risk factor-based predictive model for new-onset hypertension during pregnancy in Chinese Han women
BMC Cardiovascular Disorders (2020)
-
TRPP2 associates with STIM1 to regulate cerebral vasoconstriction and enhance high salt intake-induced hypertensive cerebrovascular spasm
Hypertension Research (2019)