Abstract
Owing to the reduction of population density and/or the environmental changes it induces, selective logging could affect the demography, reproductive biology and evolutionary potential of forest trees. This is particularly relevant in tropical forests where natural population densities can be low and isolated trees may be subject to outcross pollen limitation and/or produce low-quality selfed seeds that exhibit inbreeding depression. Comparing reproductive biology processes and genetic diversity of populations at different densities can provide indirect evidence of the potential impacts of logging. Here, we analysed patterns of genetic diversity, mating system and gene flow in three Central African populations of the self-compatible legume timber species Erythrophleum suaveolens with contrasting densities (0.11, 0.68 and 1.72 adults per ha). The comparison of inbreeding levels among cohorts suggests that selfing is detrimental as inbred individuals are eliminated between seedling and adult stages. Levels of genetic diversity, selfing rates (∼16%) and patterns of spatial genetic structure (Sp ∼0.006) were similar in all three populations. However, the extent of gene dispersal differed markedly among populations: the average distance of pollen dispersal increased with decreasing density (from 200 m in the high-density population to 1000 m in the low-density one). Overall, our results suggest that the reproductive biology and genetic diversity of the species are not affected by current logging practices. However, further investigations need to be conducted in low-density populations to evaluate (1) whether pollen limitation may reduce seed production and (2) the regeneration potential of the species.
Similar content being viewed by others
Introduction
The population density of reproductive individuals (‘population density’ for short) is central in the context of sustainability of tree resources under selective logging practices. Harvesting implies, among other effects, a reduction of population density with potential prejudicial consequences on reproduction, regeneration and genetic diversity (Degen et al., 2006; Sebbenn et al., 2008; Wernsdörfer et al., 2011; de Lacerda et al., 2013). Plant reproduction, and in particular the extent of gene flow (defined here as the sharing of alleles through mating among individuals), depends on the mating system (from selfing to outcrossing) and the pollen and seed dispersal abilities (Wilcock and Neiland, 2002; Levin et al., 2003; Duminil et al., 2009). A reduction of population density may limit the amount of pollen available for outcrossing, which could reduce seed production (pollen limitation) in obligate outcrossing species and/or increase selfing rate in self-compatible hermaphrodite species (Naito et al., 2008). In the latter case, the quality of seeds would be reduced if inbreeding depression occurs, a phenomenon commonly reported in tree species (Charlesworth, 2003; Duminil et al., 2009). However, the impact of population density reduction on animal-pollinated species is highly variable as it depends on a number of factors, including the behaviour of pollinators (Karron et al., 1995; Ghazoul, 2005). In a number of species, a reduction of population density has been shown to be counterbalanced by a change in pollinator behaviour that allows higher pollen dispersal distance (Hardy et al., 2006; Carneiro et al., 2011). Another potential long-term impact of a demographic reduction is a higher rate of loss of rare alleles, reducing the population adaptive potential (Beardmore et al., 2014). Here again, this impact might be counterbalanced if effective pollen dispersal distances increase at lower density. We could imagine, however, that pollen dispersal distances are smaller below some density threshold where a disruption of pollen flow between individuals would occur because of the unavailability of mates (Forsyth, 2003; de Waal et al., 2015).
The effect of population density reduction on plant species’ reproductive biology and genetic diversity has notably been addressed by studying the impact of logging on timber tree species. Indeed, logging results in a reduction of population density. However, logging may also affect reproductive biology through the environmental changes it induces (for example, canopy openings may affect wind movements and pollinators’ population size or behaviour, cause stress or, alternatively, favour the regeneration of light-demanding species). Studies addressing the impact of logging have provided contradictory results. Most studies have demonstrated a limited impact of logging on timber species reproductive biology and genetic diversity after one event of logging only (one cutting cycle) (Cloutier et al., 2007; Silva et al., 2008; Carneiro et al., 2011). However, modelling approaches have demonstrated that multiple cycles of logging can have a detrimental impact on genetic diversity and demography of harvested tree species (Degen et al., 2006; Sebbenn et al., 2008; Wernsdörfer et al., 2011; de Lacerda et al., 2013). Overall, these studies demonstrate that each species is unique. Accordingly, sustainable forest management practices must be species specific, taking into account the complex relationship between tree density and species reproductive biology. Unfortunately, in Africa, the biology of most tropical species is poorly documented because of the large number of species and logistical difficulties in conducting fieldwork in tropical forests.
Ideally, the impact of logging on population dynamics should be evaluated by a diachronic approach characterizing patterns of gene flow and genetic diversity several years before and after logging (Cloutier et al., 2007; Silva et al., 2008; Carneiro et al., 2011). Alternatively, the impact of logging can be investigated indirectly through a synchronic approach by comparing gene flow patterns of populations with contrasting densities (Piotti et al., 2012; Inza et al., 2012; Ojeda-Camacho et al., 2013; Fageria and Rajora, 2013). Although indirect, the latter approach has the advantage of focussing on one factor (population density) that is affected by logging. In contrast, the synchronic approach should be less affected by such confounding factors when assessing the impact of population density on processes (rather than patterns) such as selfing and pollen dispersal.
Species reproductive biology can be characterized by genetic markers, using either direct or indirect approaches. Direct methods (see, for example, Marshall et al., 1998) estimate contemporary gene flow by conducting parentage analysis, whereas indirect methods estimate either contemporary pollen flow by characterizing the spatial genetic structure of pollen clouds (Robledo-Arnuncio et al., 2007) or historical gene flow by characterizing the spatial genetic structure of adults (see, for example, Vekemans and Hardy, 2004). Although direct methods are generally preferable because they do rely on fewer model assumptions than the indirect approaches (Vekemans and Hardy, 2004), they are also more difficult to set up (availability of highly polymorphic markers, difficulties associated with exhaustive sampling in natural tropical forests and so on).
We applied both indirect and direct methods to assess gene flow and mating patterns on three different populations of Erythrophleum suaveolens, an important timber tree species in Central Africa, commonly known as ‘tali’. Here we compare the reproductive biology and genetic diversity of populations that present contrasting population densities. As outlined before, this methodology allows investigation of one aspect of the potential impact of logging on population dynamics: population density reduction. We can thus expect lower genetic diversity in a low-density population if gene flow does not compensate for an increase in local genetic drift. In the present paper, we analysed whether a lower population density was associated with: (1) lower genetic diversity, (2) stronger spatial genetic structure due to higher local genetic drift, (3) higher selfing rate and inbreeding, (4) fewer pollen donors contributing to the pollination of each mother tree and (5) a change of pollen dispersal distances. Theoretically, our expectations (1) to (4) should hold if the extent of pollen dispersal is unaffected by population density, or decreases at low density, whereas an increase in pollen dispersal distances under low population density (relationship (5)) might result in a lack of evidence to support any of the other expectations.
Materials and methods
Species description
E. suaveolens (Guill. et Perr.) Brenan (syn. E. guineense G. Don.) is an important timber species (up to 40 m in height) that belongs to the Fabaceae–Caesalpinioideae. It has widespread distribution in tropical Africa, occuring east–west from Senegal to Sudan and Kenya, and southward to Mozambique and Zimbabwe (Aubréville, 1959, 1970). Within its Central Africa distribution, the species inhabits inland semi-evergreen and evergreen rainforests (Duminil et al., 2010). Individuals are hermaphrodites (Aubréville, 1970), producing racemes up to 12-cm long that bear small-size yellowish white to greenish yellow flowers. The mean flowering time of E. suaveolens individuals is ca. 2 months, whereas the mean flowering time of the population is ca. 4 months (F Feteke, Gembloux Agro-Bio Tech, unpublished results). Pollen dispersal is probably assisted by small insects as suggested by the size of the flowers and by observations in the congeneric species Erythrophleum fordii (Zhu et al., 2009). Pods do not present any morphological device for dispersal, suggesting predominant barochory. Seeds in fresh pods are surrounded by a mucilage (Guion, 2011) that might have a nutritive value leading to secondary seed dispersal by animals. This is further supported by the presence of seeds in the faeces of primates (Poulsen et al., 2001), including gorilla (Petre et al., 2015). The soil seedbank of the species is important and seeds probably remain viable for ⩾2 years (Daïnou et al., 2011). The species presents a bell-shaped distribution of diameter size classes (Supplementary Material 1 online), with little representation of low- and high-diameter classes (Kouadio et al., 2014), suggesting reduced natural regeneration in mature closed-canopy forests.
As for all rainforest timber species from Central Africa, E. suaveolens is harvested by selective logging (Ruiz Pérez et al., 2005). Logging carried out in Cameroon, Democratic Republic of Congo (DRC) and Gabon does not follow exactly the same standards (Perthuisot and Durrieu De Madron, 2008).The rate of logging is calculated taking into account: (1) a cutting cycle (20 to 30 years according to the country or the logging company); (2) a minimum recovery rate (ratio of the number of trees that will reach the minimum diameter-felling limit at the end of the cutting cycle to the number of trees that already attained the diameter-felling limit before the cutting cycle; the threshold for recovery rate per species was fixed at 40% in Gabon and DRC and 50% in Cameroon); and (3) a minimum diameter-felling limit (fixed at 70 cm diameter at breast height (DBH) in Gabon and 50 cm DBH in Cameroon and DRC).
Sampling
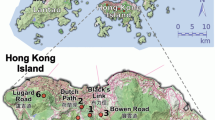
Adult trees are defined as individuals that can contribute to pollination (DBH above 30 cm according to Kouadio, 2009). Leaves or cambium of adult trees and offspring (seeds and leaves of seedlings) were collected in three different populations from Central Africa (Figure 1 and Supplementary Material 2 online). The first population is located in Cameroon within the Forest Stewardship Council (FSC)-certified ‘Pallisco’ logging concession (East province; mean coordinates: 14.34°E, 3.28°N). The second population is located in DRC within the ‘Compagnie de Transport et d’Exploitation Forestière (COTREFOR)’ logging concession (Orientale province; mean coordinates: 25.48°E, 1.00°N). The last population is located in Gabon within the FSC-certified ‘Precious Woods’ logging concession (Ogooué-Lolo province; mean coordinates: 13.03°E, 0.79°S). Sampling was done in January–March 2012 in Cameroon and Gabon and in August 2013 in DRC.
Localization of the three populations and sampling scheme in each population. Upper left: localization of the three studied populations of Erythrophleum suaveolens in the Central African forest (grey shaded area represents the potential rainforest area). Upper right: sampling scheme in Cameroon. An exhaustive sampling was conducted in a 47.5-ha plot as represented in the embedded figure. Lower left: sampling scheme in Gabon. Lower right: sampling scheme in DRC. An exhaustive sampling was conducted in a 100-ha plot as represented in the embedded figure.
These populations present contrasting tree densities (D, population density measured for DBH >30 cm), with the highest density found in Cameroon (1.72 ind ha−1), an intermediate density in DRC (0.68 ind ha−1) and the lowest density in Gabon (0.11 ind ha−1). In Cameroon and DRC we sampled a maximum number of individuals (adults, offspring) in 47.5 and 100 ha plots respectively and along three or four 4.5 to 11 km long transects departing from each plot (Figure 1). These two populations were never logged. In contrast, the population from Gabon has been logged twice, first, 15 years ago primarily targeting Aucoumea klaineana. Although E. suaveolens was not logged at that time, the opening of the canopy induced by this logging event has probably favoured the regeneration of E. suaveolens that would explain the relatively higher representation of low diameter classes in this population compared with the other two populations (Supplementary Material 1 online). Second, 60 to 70% of individuals of E. suaveolens with a DBH >70 cm were logged in the sampling zone between 2010 and 2011. This recent logging event has favoured species regeneration as attested by the high number of available seedlings (see below) in contrast to the other two populations.
In Gabon, where population density was very low, we sampled as many individuals as possible in a zone of ∼7000 ha. In total 177, 88 and 31 adult trees were sampled respectively in Cameroon, DRC and Gabon, among which 72, 33 and 23 were mother trees from which offspring families were also sampled. Seeds or seedlings’ leaves were collected on the ground below mother trees (Supplementary Material 2 online). Seeds were predominantly available in Cameroon and DRC, whereas seedlings were predominantly available in Gabon. Most progeny families were composed of seven or eight offspring (Supplementary Material 3 online). Note that this difference in sampling (seeds versus seedlings) could have consequences for the estimation of selfing rate if an elimination of selfed individuals occurs between the seed and seedlings stage.
Genetic diversity and consanguinity characterization
DNA extraction and genotyping of nine microsatellite markers were carried out as described in Duminil et al. (2011). Using SPAGeDi v.1-5 (Hardy and Vekemans, 2002), we estimated for each population and cohort (adults, seedlings, seeds) (1) the effective number of alleles (NAE) following Nielsen et al. (2003); (2) the allelic richness expressed as the expected number of alleles among k gene copies (AR(k=24)); (3) the expected heterozygosity (gene diversity) corrected for sample size (HE) (Nei, 1978); (4) the observed heterozygosity (HO); and (5) the inbreeding coefficient (FIS).
Differences in genetic diversity parameters between cohorts and populations were tested using an analysis of variance procedure in R (R Core Team, 2013). The different genetic parameters (NAE, AR, HE, HO, FIS) were estimated for each locus, cohort and population. The parameters were compared among populations for adults only accounting for the locus effect. Then, for each population, the parameters were compared among cohorts accounting for the locus effect.
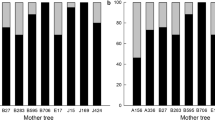
The presence of null alleles has previously been demonstrated (Duminil et al., 2011). As null alleles cause a bias in the proportion of heterozygotes used to estimate Wright’s inbreeding coefficient F, we also used INEST (Chybicki and Burczyk, 2009) under a Population Inbreeding Model to estimate for each cohort and population F(null), an estimator of inbreeding coefficient that removes the bias caused by null alleles. In each population we tested whether the level of inbreeding was significantly different between cohorts by applying unpaired t-tests on (H0) per individual (proportion of heterozygous loci) in each cohort, considering only individuals genotyped for at least seven out of nine loci. Under inbreeding depression, if inbred (that is, less heterozygous) individuals are counter selected at early life stages, we expect to observe more heterozygosity in adults than in seeds and/or seedlings.
Fine-scale spatial genetic structure
Spatial genetic structure (SGS) of adult trees was assessed for each population. Pairwise kinship coefficients (Fij) between individuals (Loiselle et al., 1995) and 95% confidence intervals were estimated at regular geographical distance intervals using SPAGeDi v.1-4c (Hardy and Vekemans, 2002). SGS was tested by permuting randomly the position of the individuals (10 000 randomizations). Estimates of the Sp statistic (a synthetic measure of SGS strength) were obtained for each population from the slope of the regression of pairwise kinship coefficient on ln(distance) and the mean pairwise kinship coefficient measured at the first distance class (F1), following Vekemans and Hardy (2004).
Additionally, assuming drift–dispersal equilibrium, we estimated the neighbourhood size and the gene dispersal distance σg for each population relying on SGS patterns following the procedure described in Hardy et al. (2006). The principle of the method is that Fij is expected to decay linearly with the ln(distance) at a rate inversely proportional to the product DE.σg2, at least for a distance range between σg and ca. 20 σg, where DE is the effective density of reproductive individuals and σg2 is the axial variance of gene dispersal distance between two generations. As the regression must be performed on a distance interval depending on the parameter to estimate (σg), an iterative procedure is implemented in SPAGeDi and should converge only if data were sampled at an adequate spatial scale (Hardy et al., 2006). Different values of DE were tested, considering 1/2, 1/4 and 1/8 of the adult densities to account for the lifetime variation in reproductive success among adult trees (Hardy et al., 2006). Thus, we tested respectively for Cameroon, DRC and Gabon the following effective densities: 0.850/0.425/0.212, 0.340/0.170/0.085 and 0.054/0.027/0.013 (ind ha−1). Approximate standard errors are obtained by jackknifing over loci.
Assignments of offspring to mothers
As a prerequisite to mating system and pollen pool analyses, given the potential for secondary seed dispersal, we first tested whether the genotypes of candidate mothers and seeds/seedlings that were collected below them were compatible with a mother–offspring relationship. We conducted a maternal analysis using CERVUS (Marshall et al., 1998). CERVUS uses a maximum likelihood approach and assigns maternity according to the highest likelihood (LOD score). Simulations were conducted to estimate the critical values of LOD score required to assign maternity with a given degree of confidence (80 and 95% confidence levels). The following simulation parameters were applied to define the confidence level of maternity analysis assignment: 10 000 simulated mating events; all adults in a population were considered to be candidate mothers; individuals were typed at a minimum of five loci; 90% of candidate mothers were sampled; and a genotyping error rate of 0.1. Only offspring correctly assigned to the expected mother (mother localized above collected offspring) were used in the following steps. When seeds or seedlings were not assigned to the expected mothers, presumably because of secondary dispersal, we did not tentatively reassign them.
Mating system analyses
For each population, we estimated the outcrossing rate (t) in four ways. First, we can rely on F(null) seeds (Cameroon and DRC) or F(null) seedlings (Gabon) obtained from INEST to estimate the outcrossing rate. The outcrossing rate can be calculated from F(null) through the relation t=(1−F(null))/(1+F(null)) (Fyfe and Bailey, 1951) that assumes that inbreeding results only from selfing, there is no inbreeding depression and that the inbreeding of adults is at equilibrium. However, as we detected strong inbreeding depression so that adults were non-inbred (see below), we used the relation t=1−2.F(null) that assumes they are in Hardy–Weinberg equilibrium despite selfing because seeds or seedlings resulting from selfing never reach the adult stage.
Second, outcrossing rate per population was also estimated by leading paternity analyses in CERVUS (Marshall et al., 1998). Assigned mothers (see maternity analysis above) were fixed for each offspring and the paternity analysis was conducted using the self-fertilization option. The following simulation parameters were applied to define the confidence level of paternity analysis assignment: 10 000 simulated mating events; all adults of the given population as candidate father plants; individuals typed at a minimum of five loci; 0.5 as the proportion of candidate fathers sampled; genotyping error rate of 0.1. The outcrossing rate was estimated as the number of observed outcrossing events over the total number of tested offspring.
The per-family self-fertilization rate was estimated for each individual of each family using a Bayesian approach implemented in the MSF software (Chybicki, 2013a, 2013b). The method is based on the mixed mating model, with rates of self-fertilization treated separately for each maternal individual. Markov Chain Monte Carlo analyses were run for 1 million cycles with sampling of the parameters every 1000 steps. The per-family selfing rate was then estimated using a burn-in of 10%. The difference of these selfing rates between populations was tested using unpaired t-tests.
Finally, the multilocus outcrossing rate (tm) and the single-locus outcrossing rate (ts) were estimated using MLTR v.3.2 (Ritland, 2002) from progeny arrays (N=58 families for Cameroon, 32 for DRC and 23 for Gabon). Mating among relatives (biparental inbreeding) was estimated by the difference (tm−ts). In case of biparental inbreeding, it is expected that (ts)<(tm), and the difference is a minimum estimate of the apparent selfing because of biparental inbreeding (Shaw et al., 1981). Standard deviation of these estimators was evaluated through a bootstrap procedure (1000 repetitions).
Spatial structure of pollen pools
Relying on mapped mother–offspring genotypic data, contemporary pollen dispersal was inferred using the so-called KINDIST and TWOGENER methods as implemented in the software POLDISP (Robledo-Arnuncio et al., 2007). We followed recommendations from POLDISP’s user manual in the preparation of the data sets per population: no mismatch between mother and offspring (loci presenting mismatches were transformed as missing data in offspring), no missing data for the mothers, no seeds resulting from selfing (we used the results from the CERVUS analysis to removed selfed seeds) and a minimum of two offspring per mother. We first used KINDIST to estimate the correlation of paternity of outcrossed progenies within families and between maternal families (separately for each pair of families) from the mapped genotypes of mother–offspring data. The mean number of effective pollen donors (Nep) that participate to cross-pollination was estimated from the within-sibship correlated paternity (rp) as Nep=1/rp. For comparison (rp) was also calculated with MLTR (Ritland, 2002) using as input files all correctly-assigned-to-mother offspring (including those resulting from selfing).
To test whether the among-sibship correlated paternity was inversely correlated with the distance between mother trees, we used a Mantel test procedure as implemented in the zt software (Van de Peer, 2002). The slope was negative and significant in each population. We then tested the fit of our data with the different pollen dispersal distribution models available in POLDISP (normal, exponential, exponential power). We used 1000 m (Cameroon and DRC) and 2000 m (Gabon) as reference threshold distances to define unrelated pollen pools, because there was no decrease of the among-sibship correlated paternity beyond these threshold distances. For each population, the best dispersal distribution was chosen by comparing the least-square residuals obtained for each of these distributions. Finally, we used TWOGENER to estimate the effective male population density (DEp) having as input the pollen dispersal distribution parameters estimated with KINDIST. The ratio (DEp/D) provides an indication of the proportion of adult trees that have contributed to reproduction as pollen donors within the population for one given year.
Results
Genetic diversity and consanguinity
None of the genetic diversity parameters calculated for the adult cohort differed significantly among populations (data not shown). Only the population in Gabon presented differences in genetic diversity among cohorts (for NAE, AR and HE; see Table 1), but this can be attributed to a sampling effect (low representation of the seed cohort with 23 seeds coming from only 4 mother trees). The (uncorrected) inbreeding coefficient (F) was significantly higher than zero in all populations and cohorts. Estimates correcting for the presence of null alleles, F(null), showed a decreasing trend from seeds to adults: F was slightly lower in seedlings than in seeds and strongly decreased from seedlings to adult stage. In adults, F(null) never departed significantly from zero, indicating that they are not inbred. The unpaired t-test on observed heterozygosity per individual was significant (P<0.05) only between adults and seeds in Cameroon and DRC, and between adults and seedlings in Cameroon. The nonsignificant result in Gabon between adults and seedlings may be due to the low sample size of adult individuals in this population because mean heterozygosity values showed the same trends as in the other populations.
Fine-scale SGS and inference of gene dispersal distances
In all three populations a signal of isolation by distance was observed with pairwise kinship coefficients decreasing significantly with distance (Figure 2 and Table 2). In all populations, kinship for the first distance class (ca. 100 m for Cameroon and DRC, ca. 500 m for Gabon) ranged from 0.04 to 0.06, and quickly dropped with distance, indicating that spatially close individuals are more related. The kinship-distance curve for the low-density Gabonese population was similar to the ones of the medium- to high-density populations but shifted towards larger distances (Figure 2), whereas the Sp statistics, which quantify the strength of SGS, were similar for all three populations, ranging from 0.0053 to 0.0073 (Table 2). The procedure to estimate gene dispersal parameters converged only for the Cameroonian population. For DE=0.820 ind ha−1, σg=379±67 m (mean±s.e.), and neighbourhood size Nb=148±54. For DE=0.410 ind ha−1, σg=483±64 m, and neighbourhood size Nb=120±33. No convergence was reached for DE=0.205 ind ha−1.
Average kinship coefficients Fij between pairs of individuals at different geographical distance intervals (log scale) in each population. Vertical bars are s.e.m. The continuous grey line, the continuous black line and the dashed grey line correspond respectively to the Cameroonian, the Congolese and the Gabonese populations.
Assignment of offspring to mothers
For the Cameroonian population, 384 out of 499 seeds (∼77%) were assigned to the expected mother. In this population, we did not tentatively assign collected seedlings to mothers. For the Congolese and Gabonese populations, respectively 200 out of 254 seeds and seedlings (∼79%), and 179 out of 199 seeds and seedlings (90%) were assigned to the expected mother.
Mating system analyses
All three populations of E. suaveolens presented a mixed mating system (mixtures of selfed and outcrossed pollination). We found a linear trend between selfing rate and population density with one of the methods used (MSF software, Table 2), which was contrary to our expectations as selfing increases with increasing population density. However, the unpaired t-test between populations (based on the per-family self-fertilization rates) demonstrates that estimates of outcrossing rates are only significantly different between the Gabonese (tm=0.850) and Cameroonian (tm=0.782) populations. This is potentially only because of a methodological bias as the selfing rate could have been underestimated in the population from Gabon where seedlings had to be used instead of seeds. The Gabonese population also presented a higher biparental inbreeding signal as measured by (tm−ts) than the two other populations.
Spatial structure of pollen pools
In all three populations, the among-sibship correlated paternity was negatively and significantly correlated with distance between mother trees (Supplementary Material 4 online). MLTR and POLDISP provided similar estimates of correlated paternity within maternal sibships (rp) for each population (Table 3). The differences between populations were not significant (P>0.05 for all comparisons, unpaired t-test). Despite a more than twofold higher population density (D) in Cameroon than in DRC, this latter population presented only a slightly lower proportion of individuals participating in pollination (D/DEp) (Table 3). However, the number of effective pollen donors (NEP) was higher in the Congolese population than in the Cameroonian one. The Gabonese population presented the lowest DE and NEP. In the absence of confidence intervals around these measures, we were not able to test whether differences were statistically significant. The best fit of the dispersal distribution was obtained with the exponential dispersal kernel in all three populations. The average pollen dispersal distance estimates decreased from 1001 m in the low-density population to 195 m in the high-density population (Table 3).
Discussion
We analysed patterns of genetic diversity and structure, mating system and gene flow in three populations of the African timber tree species E. suaveolens. The three populations present different population densities (for trees with DBH >30 cm). The study of the relationship between contrasting population densities and reproductive biology or genetic diversity parameters can be used to predict one aspect of the potential impact of logging. The Cameroonian population presented the highest density followed by the population from DRC (ca. 40% of the Cameroonian one; Table 3) and finally the population from East Gabon with the lowest density (ca. 6% of the Cameroonian one). Importantly, these results concern only E. suaveolens and not E. ivorense, both commonly named as tali, but that actually correspond to two clearly differentiated parapatric species (Duminil et al., 2010).
Lower population density is not associated with lower genetic diversity
No relationship between genetic diversity and population density is supported by our results. Levels of genetic diversity are equivalent across the three populations and across cohorts within each population (Table 1). A previous study indicated that the Cameroonian and the Gabonese populations are located in two different gene pools with equivalent levels of genetic diversity (Duminil et al., 2013). Our data, acquired at a fine-geographical scale, support this result, highlighting that historical factors did not lead to different levels of genetic diversity. Estimates of genetic diversity of the seed cohort also depend on pollen dispersal distance (pollen diversity). The larger the pollen distance, the greater the degree seeds will ‘capture’ the pollen diversity of surrounding individuals. We have demonstrated that pollen dispersal distance is inversely related to population density. This indicates that the absence of relationships between population density and genetic diversity in the case of E. suaveolens actually corresponds to a drift/migration balance: local genetic drift would be expected to be higher in the low-density population (the strength of drift is inversely proportional to population size), but it is prevented by long-distance pollen dispersal (see below), maintaining genetic diversity.
Nevertheless, we need to be careful about extrapolation of such a conclusion. Despite the absence of a relationship between genetic diversity and population density in this study and others (Cloutier et al., 2007; Fageria and Rajora, 2013), it is not certain that genetic diversity is secure. Indeed, long-term impacts of selective logging across multiple cutting cycles investigated through modelling have led to detrimental effects (Degen et al., 2006; Sebbenn et al., 2008; Wernsdörfer et al., 2011; but see Vinson et al., 2014). One of these models particularly emphasized the impact of juvenile mortality on genetic diversity (Wernsdörfer et al., 2011). We do not have specific data on E. suaveolens juvenile mortality, but the population structure is clearly truncated at small-diameter classes in the two nonlogged populations (Cameroon and DRC; Supplementary Material 1 online), indicating the poor regeneration capacity of the species under closed canopy as expected for light-demanding species. This suggests that the future replacement of seed trees after logging is not ensured at the same level, unless intensive enrichment planting is carried out as done by some FSC-certified logging companies of Central Africa. This effect has to be carefully modelled before drawing any clear conclusion on the long-term impact of logging on E. suaveolens genetic diversity. Further information on the impact of cutting on species’ regeneration is needed to propose recommendations. E. suaveolens is a light-demanding species in its early stages and opening the canopy by logging might have a positive influence on its regeneration (Kouadio, 2009) if the openings are large enough. Moreover, important parameters such as cutting diameter, growth rates, natural mortality rate and time between two cutting cycles have to be integrated for accurate interpretation of results.
Lower population density is not associated with stronger spatial genetic structure
The strength of SGS depends on a migration–drift equilibrium. Overall, the strength of E. suaveolens SGS as measured by the Sp statistic is typical of tree species (Table 2; Vekemans and Hardy, 2004) and suggests that seed-mediated dispersal is relatively efficient in this species (Dick et al., 2007). It is expected that lower-density populations present stronger SGS as local drift would be more important (Vekemans and Hardy, 2004). Here, we can only test this relationship between the population from Cameroon and the population from DRC as the SGS estimation for the population from Gabon lacks precision because of a small sample size. The population from DRC does not seem to present a stronger SGS than the population from Cameroon, suggesting that the strength of SGS is not related to population density (large overlap of s.e.m. values; Table 2). This suggests that the potentially larger drift effect expected at lower densities is compensated by increased dispersal distance (see below). Such a pattern has already been observed in another African tropical tree species, Aucoumea klaineana (Born et al., 2008).
Lower population density is not associated with higher selfing rate and inbreeding
There is no clear relationship between population density and mating system at least for the range of densities observed in the current study (Table 2). The only significant difference, between the lowest and the highest population density (between Gabon and Cameroon) is potentially only due to a methodological bias. Altogether, all three populations seem to present similar levels of selfing, regardless of population density (between 13 and 19% based on MLTR analyses; Table 3). The same result has already been obtained for a set of timber species, where the selfing rate has been demonstrated to remain unchanged after selective logging (Cloutier et al., 2007; Lourmas et al., 2007; Ojeda-Camacho et al., 2013). However, in these studies, the outcrossing rate was close to one that tends to support the existence of a strong auto-incompatibility system. Here, we also observed similar levels of selfing despite different population densities. The stability of this mixed mating system is puzzling. It has been proposed that the evolutionary stability of mixed mating systems is determined by the timing and relative amount of self- and outcross-pollination (Holsinger, 1991). In other words, if outcross pollen supply is limited, the selfing rate will be higher. We can expect higher pollen limitation in low-density populations, but this is probably not the case for E. suaveolens, as pollen distance increases with decreasing density (see below). The compensation of population density reduction by larger pollen flow would explain that selfing rate is relatively stable in populations that present different levels of density. Alternatively, the relative stability of the mixed mating system could be explained by post-pollination mechanisms, such as partial self-incompatibility systems (Goodwillie et al., 2005).
Like most tree species, E. suaveolens is subject to inbreeding depression. Indeed, we observed a trend of decreasing inbreeding from seed to adult stage (Table 1), suggesting that individuals resulting from selfing die before reaching the adult stage (Barrett and Charlesworth, 1991). This trend was significant only for two out of the three populations (Cameroon and DRC). Selfing is generally detrimental in tree species because of their sensitivity to inbreeding depression, a likely consequence of their long generation time (Charlesworth, 2003; Duminil et al., 2009). This expression of inbreeding depression has important consequences in terms of species population dynamics in relation to selective logging. Adult individuals that become genetically isolated after logging will have a very low probability of producing viable descendants. Though we have demonstrated that selfing rate is somewhat independent from population density, studying the relationships between degree of genetic isolation associated with greater distances than observed in the current study and mating system/seed set of trees in low-density populations of E. suaveolens will be necessary to further test the existence of this detrimental effect.
Biparental inbreeding seems to be more pronounced in the low-density population than in the high-density one (Table 2). The main cause of biparental inbreeding might be the low efficiency of seed-mediated gene flow as we demonstrated that pollen-mediated gene flow decreases with population density. If distance of seed dispersal is similar (and low) in all three populations, it is expected that the lowest density population is also the one most prone to biparental inbreeding. Here we did not find any clear relationship between population density and level of SGS, which tends to contradict this hypothesis. However, the absence of an accurate estimation of SGS for the low-density population due to small sample size limits our capacity for interpretation. Alternatively, the presence of a biparental inbreeding signal could suggest preferential mating between related individuals as would be the case if phenological asynchrony has a genetic origin. In the absence of phenological data, we cannot further interpret our results.
Lower population density is not associated with fewer pollen donors contributing to the pollination of each mother tree
Levels of correlated paternity within maternal sibships are similar in all three populations (Table 3). This result is well in line with an increase in the average distance of pollen dispersal with decreasing density and further provides information on the quality of mating events (similar number of pollen donors in different population densities). One can expect that this qualitative estimation can also indicate the quantity of pollen received, but this is actually not necessarily the case. The estimations that have been obtained in this study provide a picture of mating events based only on collected seeds or seedlings. These observations are independent from the actual level of seed production. We can obtain similar patterns of correlated paternity in the presence or in the absence of pollen limitation (with respectively low and high seed production). Measures of seed production through direct observations at different classes of physical isolation to the nearest neighbour need to be obtained to get a clearer view of the quantitative aspects of mating events.
Our results suggest that the effective density of pollen donors might be more reduced in low-density populations than in the high-density ones. This result needs to be interpreted with caution as we were not able to test the statistical significance of this trend. This result is, however, in line with our expectation and with other studies (Mimura and Aitken, 2007; Tani et al., 2012; Masuda et al., 2013).
Lower population density is associated with a change of pollen dispersal distances
The extent of gene flow is correlated with population densities as pollen dispersal distances have been shown to decrease with population density. The average distance of pollen dispersal varies from 200 m (Cameroon) to 1000 m (Gabon; Table 3). In other words, these results suggest that at lower population densities, trees are still connected as a result of larger pollen dispersal distances. If pollinating species are identical in all three populations, this can outline their capacity to travel longer distances when flowering trees are more distant. Increased pollen dispersal distances in the case of lower tree densities have already been reported for other tropical species (White et al., 2002; Dick et al., 2003; Born et al., 2008). This pattern probably explains the absence of relationships between population density and previously discussed factors (genetic diversity, SGS, mating system, inbreeding depression) within E. suaveolens.
Logging implications
Norms differ among the three studied countries on the minimum cutting diameters. Logging intensity of E. suaveolens is theoretically lower in Gabon (minimum cutting diameter of 70 cm) than in Cameroon and DRC (minimum cutting diameter of 50 cm). Our results suggest that such logging intensities in populations with the same characteristics as those studied in Cameroon and DRC would not affect a population’s viability in a single cutting cycle. Given logging norms in these countries, we can estimate that population density of adult individuals would be reduced to 0.47 and 0.30/ha respectively in Cameroon and DRC. Such population densities are still higher than the population density in Gabon. However, models need to be developed to investigate the impact of multiple cutting cycles. It is difficult to predict the impact of logging on the population from Gabon as we have no indication of pollen dispersal distances at lower densities. This study needs to be completed by additional data on pollen limitation, notably through the investigation of the relationships of seed set, fruit phenology, seed–tree distance to the nearest neighbour and selfing rate. We recommend conducting such a study in populations that present densities equivalent to or lower than the population from Gabon. Such studies require exhaustive sampling, detailed cartography of the population structure and long-term monitoring of the flowering and fruiting phenology, which are challenging to organize in the field.
Conclusion
The comparison of three different populations with contrasting densities of the timber species E. suaveolens provides indications of the relationships between population density, mating pattern and genetic diversity. Equivalent levels of genetic diversity, selfing rates and correlated paternity suggest that these population parameters are resilient to a decrease of population density, one of the key consequences of selective logging. In particular, although the species exhibits inbreeding depression, there is no evidence of a loss of quality of the seed produced at low densities. This stability can be explained by the negative relationship observed between pollen dispersal distances and population density, a phenomenon that may be prevalent in plant populations. Although these results are reassuring regarding the risk that selective logging may impede the regeneration of timber tree populations, our study did not allow us to test whether seed production was affected by a decrease in density, nor to address all the other consequences of logging, notably those related to habitat perturbations, which were not the focus of the study. Additional work is thus necessary to provide logging recommendations, such as the definition of a maximum distance to be kept between seed trees to allow the qualitative and quantitative reproduction of the species. We suggest that investigations need to be conducted in low-density populations, which are more threatened by population reduction.
Data Archiving
Genotype data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.b78fb.
References
Aubréville A . (1959). La flore forestière de la Côte d’Ivoire, Tome 1. Nogent-sur-Marne, Cent Tech For Trop (Ctft) 1: 370.
Aubréville A . (1970) Légumineuses-Caesalpinioidées. In: Aubréville A, Leroy J-F (eds). Flore du Cameroun 9, Muséum national d’Histoire naturelle: Paris. pp 1–339.
Barrett SCH, Charlesworth D . (1991). Effects of a change in the level of inbreeding on the genetic load. Nature 352: 522–524.
Beardmore T, Loo J, Mathews S, McGovern K, Nikiema A, Souvannavong O et al. (2014) Value and importance of forest genetic resources In The State of the World’s Forest Genetic Resources. FAO - Commission on Genetic Resources for Food and Agriculture: Rome, Italy. pp 19–26.
Born C, Hardy OJ, Chevallier MH, Ossari S, Atteke C, Wickings J et al. (2008). Small-scale spatial genetic structure in the Central African rainforest tree species Aucoumea klaineana: a stepwise approach to infer the impact of limited gene dispersal, population history and habitat fragmentation. Mol Ecol 17: 2041–2050.
Carneiro FS, Lacerda AEB, Lemes MR, Gribel R, Kanashiro M, Wadt LHO et al. (2011). Effects of selective logging on the mating system and pollen dispersal of Hymenaea courbaril L. (Leguminosae) in the Eastern Brazilian Amazon as revealed by microsatellite analysis. For Ecol Manage 262: 1758–1765.
Charlesworth D . (2003). Effects of inbreeding on the genetic diversity of populations. Philos Trans R Soc London Ser B Biol Sci 358: 1051–1070.
Chybicki IJ . (2013a) MSF 1.01 (Mating System with the F-model). Sofware manual.
Chybicki IJ . (2013b). Note on the applicability of the F-model in analysis of pollen pool heterogeneity. J Hered 104: 578–585.
Chybicki IJ, Burczyk J . (2009). Simultaneous estimation of null alleles and inbreeding coefficients. J Hered 100: 106–113.
Cloutier D, Kanashiro M, Ciampi AY, Schoen DJ . (2007). Impact of selective logging on inbreeding and gene dispersal in an Amazonian tree population of Carapa guianensis Aubl. Mol Ecol 16: 797–809.
Daïnou K, Bauduin A, Bourland N, Gillet J-F, Fétéké F, Doucet J-L . (2011). Soil seed bank characteristics in Cameroonian rainforests and implications for post-logging forest recovery. Ecol Eng 37: 1499–1506.
Degen B, Blanc L, Caron H, Maggia L, Kremer A, Gourlet-Fleury S . (2006). Impact of selective logging on genetic composition and demographic structure of four tropical tree species. Biol Conserv 131: 386–401.
De Lacerda AEB, Roberta Nimmo E, Sebbenn AM . (2013). Modeling the long-term impacts of logging on genetic diversity and demography of Hymenaea courbaril. For Sci 59: 15–26.
de Waal C, Anderson B, Ellis AG . (2015). Relative density and dispersion pattern of two southern African Asteraceae affect fecundity through heterospecific interference and mate availability, not pollinator visitation rate. J Ecol 103: 513–525.
Dick CW, Etchelecu G, Austerlitz F . (2003). Pollen dispersal of tropical trees (Dinizia excelsa: Fabaceae) by native insects and African honeybees in pristine and fragmented Amazonian rainforest. Mol Ecol 12: 753–764.
Dick CW, Hardy OJ, Jones FA, Petit RJ . (2007). Spatial scales of pollen and seed-mediated gene flow in tropical rain forest trees. Trop Plant Biol 1: 20–33.
Duminil J, Brown RP, Ewédjè E-EBK, Mardulyn P, Doucet J-L, Hardy OJ . (2013). Large-scale pattern of genetic differentiation within African rainforest trees: insights on the roles of ecological gradients and past climate changes on the evolution of Erythrophleum spp (Fabaceae). BMC Evol Biol 13: 195.
Duminil J, Hardy OJ, Petit RJ . (2009). Plant traits correlated with generation time directly affect inbreeding depression and mating system and indirectly genetic structure. BMC Evol Biol 9: 177.
Duminil J, Heuertz M, Doucet JL, Bourland N, Cruaud C, Gavory F et al. (2010). CpDNA-based species identification and phylogeography: application to African tropical tree species. Mol Ecol 19: 5469–5483.
Duminil J, Koffi KG, Debout GDG, Sebastiani F, Vendramin GG, Heuertz M et al. (2011). Isolation of SSR markers for two African tropical tree species Erythrophleum suaveolens and E. ivorense (Caesalpinioideae). Am J Bot Prim Notes Protoc 98: E106–E108.
Fageria MS, Rajora OP . (2013). Effects of harvesting of increasing intensities on genetic diversity and population structure of white spruce. Evol Appl 6: 778–794.
Forsyth SA . (2003). Density-dependent seed set in the Haleakala silversword: evidence for an Allee effect. Oecologia 136: 551–557.
Fyfe JL, Bailey NTJ . (1951). Plant breeding studies in leguminous forage crops. I. Natural cross-breeding in winter beans. J Agric Sci 41: 371–378.
Ghazoul J . (2005). Pollen and seed dispersal among dispersed plants. Biol Rev 80: 413.
Goodwillie C, Kalisz S, Eckert CG . (2005). The evolutionary enigma of mixed mating systems in plants: Occurrence, theoretical explanations, and empirical evidence. Annu Rev Ecol Evol Syst 36: 47–79.
Guion H . (2011). Etude de l'écologie d'Erythrophleum suaveolens (Guill. & Perr.) Brenan (tali) et de l'origine de ses peuplements au sud-est du Cameroun. Master thesis, GxABT-ULg, p 80.
Hardy OJ, Maggia L, Bandou E, Breyne P, Caron H, Chevallier MH et al. (2006). Fine-scale genetic structure and gene dispersal inferences in 10 neotropical tree species. Mol Ecol 15: 559–571.
Hardy OJ, Vekemans X . (2002). SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620.
Holsinger KE . (1991). Mass-action models of plant mating systems: the evolutionary stability of mixed mating systems. Am Nat 138: 602–622.
Inza MV, Zelener N, Fornes L, Gallo LA . (2012). Effect of latitudinal gradient and impact of logging on genetic diversity of Cedrela lilloi along the Argentine Yungas Rainforest. Ecol Evol 2: 2722–2736.
Karron JD, Thumser NN, Tucker R, Hessenauer AJ . (1995). The influence of population density on outcrossing rates in Mimulus ringens. Heredity (Edinb) 75: 175–180.
Kouadio L . (2009) Mesures sylvicoles en vue d’améliorer la gestion des populations d'essences forestières commerciales de l'Est du Cameroun. PhD dissertation, Faculté Universitaire des Sciences Agronomiques de Gembloux: Gembloux, Belgium.
Kouadio YL, Neuba DF, Koné M, Bourland N, Doucet J . (2014). Étude de la croissance diamétrique de quatre essences de bois d’oeuvre exploitées à l’Est du Cameroun. J Appl Biosci 77: 6444–6455.
Levin SA, Muller-Landau HC, Nathan R, Chave J . (2003). The ecology and evolution of seed dispersal: a theoretical perspective. Annu Rev Ecol Evol Syst 34: 575–604.
Loiselle BA, Sork VL, Nason J, Graham C . (1995). Spatial genetic-structure of a tropical understory shrub, Psychotria Officinalis (Rubiaceae). Am J Bot 82: 1420–1425.
Lourmas M, Kjellberg F, Dessard H, Joly HI, Chevallier M-H . (2007). Reduced density due to logging and its consequences on mating system and pollen flow in the African mahogany Entandrophragma cylindricum. Heredity (Edinb) 99: 151–160.
Marshall TC, Slate J, Kruuk LEB, Pemberton JM . (1998). Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7: 639–655.
Masuda S, Tani N, Ueno S, Lee SL, Muhammad N, Kondo T et al. (2013). Non-density dependent pollen dispersal of Shorea maxwelliana (Dipterocarpaceae) revealed by a Bayesian mating model based on paternity analysis in two synchronized flowering seasons. PLoS One 8: e82039.
Mimura M, Aitken SN . (2007). Increased selfing and decreased effective pollen donor number in peripheral relative to central populations in Picea sitchensis (Pinaceae). Am J Bot 94: 991–998.
Naito Y, Kanzaki M, Iwata H, Obayashi K, Lee SL, Muhammad N et al. (2008). Density-dependent selfing and its effects on seed performance in a tropical canopy tree species, Shorea acuminata (Dipterocarpaceae). For Ecol Manage 256: 375–383.
Nei M . (1978). Estimation of average heterozygosity and genetic distance for small number of individuals. Genetics 89: 583–590.
Nielsen R, Tarpy DR, Kern Reeve H . (2003). Estimating effective paternity number in social insects and the effective number of alleles in a population. Mol Ecol 12: 3157–3164.
Ojeda-Camacho M, Kjær ED, Philipp M . (2013). Population genetics of Guibourtia chodatiana (Hassl.) J. Leonard, in a dry Chiquitano forest of Bolivia. For Ecol Manage 289: 525–534.
Perthuisot N, Durrieu De Madron L . (2008) Etude comparative des législations forestières des pays du bassin du Congo. Rapport d’étude COMIFAC. TEREA, Libreville.
Petre C-A, Tagg N, Beudels-Jamar R, Haurez B, Salah M, Spetschinsky V et al. (2015). Quantity and spatial distribution of seeds dispersed by a western lowland gorilla population in south-east Cameroon. J Trop Ecol 31: 1–12.
Piotti A, Leonardi S, Buiteveld J, Geburek T, Gerber S, Kramer K et al. (2012). Comparison of pollen gene flow among four European beech (Fagus sylvatica L.) populations characterized by different management regimes. Heredity (Edinb) 108: 322–331.
Poulsen JR, Clarck CJ, Smith TB . (2001). Seed dispersal by a diurnal primate community in the Dja Reserve, Cameroon. J Trop Ecol 17: 787–808.
R Core Team. (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. Available from http://www.R-project.org/.
Ritland K . (2002). Extensions of models for the estimation of mating systems using n independent loci. Heredity (Edinb) 88: 221–228.
Robledo-Arnuncio JJ, Austerlitz F, Smouse PE . (2007). POLDISP: a software package for indirect estimation of contemporary pollen dispersal. Mol Ecol Notes 7: 763–766.
Ruiz Pérez M, Ezzine de Blas D, Nasi R, Sayer JA, Sassen M, Angoué C et al. (2005). Logging in the Congo Basin: a multi-country characterization of timber companies. For Ecol Manage 214: 221–236.
Sebbenn AM, Degen B, Azevedo VCR, Silva MB, de Lacerda AEB, Ciampi AY et al. (2008). Modelling the long-term impacts of selective logging on genetic diversity and demographic structure of four tropical tree species in the Amazon forest. For Ecol Manage 254: 335–349.
Shaw DV, Kahler AL, Allard RW . (1981). A multilocus estimator of mating system parameters in plant populations. Proc Natl Acad Sci USA 78: 1298–1302.
Silva MB, Kanashiro M, Ciampi AY, Thompson I, Sebbenn AM . (2008). Genetic effects of selective logging and pollen gene flow in a low-density population of the dioecious tropical tree Bagassa guianensis in the Brazilian Amazon. For Ecol Manage 255: 1548–1558.
Tani N, Tsumura Y, Fukasawa K, Kado T, Taguchi Y, Lee SL et al. (2012). Male fecundity and pollen dispersal in hill dipterocarps: significance of mass synchronized flowering and implications for conservation. J Ecol 100: 405–415.
Van de Peer Y . (2002). ZT: a software tool for simple and partial Mantel tests. J Stat Softw 7: 1–12.
Vekemans X, Hardy OJ . (2004). New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13: 921–935.
Vinson CC, Kanashiro M, Sebbenn AM, Williams TC, Harris SA, Boshier DH . (2014). Long-term impacts of selective logging on two Amazonian tree species with contrasting ecological and reproductive characteristics: inferences from Eco-gene model simulations. Heredity (Edinb) 115: 130–139.
Wernsdörfer H, Caron H, Gerber S, Cornu G, Rossi V, Mortier F et al. (2011). Relationships between demography and gene flow and their importance for the conservation of tree populations in tropical forests under selective felling regimes. Conserv Genet 12: 15–29.
White GM, Boshier DH, Powell W . (2002). Increased pollen flow counteracts fragmentation in a tropical dry forest: an example from Swietenia humilis Zuccarini. Proc Natl Acad Sci USA 99: 2038–2042.
Wilcock C, Neiland R . (2002). Pollination failure in plants: why it happens and when it matters. Trends Plant Sci 7: 270–277.
Zhu P, Ye WH, Wang ZF, Cao HL, Zhang M, Li L et al. (2009). Isolation and characterization of ten polymorphic microsatellite in the endangered tree Erythrophleum fordii oliv. Conserv Genet 10: 1017–1019.
Acknowledgements
Research was supported in part by the Congo Basin Forest Fund (Beyond Timber project; P-Z1-C00-024), the CGIAR Research Program on Forests, Trees and Agroforestry and the Belgian Science Policy (Belspo) through project AFRIFORD. We are grateful to the logging companies Pallisco, Precious Woods and Compagnie de Transport et d’Exploitation Forestière (COTREFOR) for providing access to their concessions and technical support. We also thank Nature+asbl, Millet and Pasquet groups for financial and technical support. We acknowledge the field teams especially Paul Lagoute, Patrice Tekam, Crépin Ndjopamdé, Théophile Ayol and Armand Boubady. We thank several anonymous referees for helpful comments on earlier drafts of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Duminil, J., Daïnou, K., Kaviriri, D. et al. Relationships between population density, fine-scale genetic structure, mating system and pollen dispersal in a timber tree from African rainforests. Heredity 116, 295–303 (2016). https://doi.org/10.1038/hdy.2015.101
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2015.101
This article is cited by
-
Stand development stages and recruitment patterns influence fine-scale spatial genetic structure in two Patagonian Nothofagus species
Annals of Forest Science (2022)
-
Population genetic structure and connectivity of a riparian selfing herb Caulokaempferia coenobialis at a fine-scale geographic level in subtropical monsoon forest
BMC Plant Biology (2021)
-
Evidence of past forest fragmentation in the Congo Basin from the phylogeography of a shade-tolerant tree with limited seed dispersal: Scorodophloeus zenkeri (Fabaceae, Detarioideae)
BMC Ecology and Evolution (2021)
-
Seed and pollen dispersal and fine-scale spatial genetic structure of a threatened tree species: Pericopsis elata (HARMS) Meeuwen (Fabaceae)
Tree Genetics & Genomes (2021)
-
Short distance pollen dispersal and low genetic diversity in a subcanopy tropical rainforest tree, Fontainea picrosperma (Euphorbiaceae)
Heredity (2019)