Abstract
The risks of gene flow between interfertile native and introduced plant populations are greatest when there is no spatial isolation of pollen clouds and phenological patterns overlap completely. Moreover, invasion probabilities are further increased if introduced populations are capable of producing seeds by selfing. Here we investigated the mating system and patterns of pollen-mediated gene flow among populations of native ash (Fraxinus excelsior) and mixed plantations of non-native ash (F. angustifolia and F. excelsior) as well as hybrid ash (F. excelsior × F. angustifolia) in Ireland. We analysed the flowering phenology of the mother trees and genotyped with six microsatellite loci in progeny arrays from 132 native and plantation trees (1493 seeds) and 444 potential parents. Paternity analyses suggested that plantation and native trees were pollinated by both native and introduced trees. No signs of significant selfing in the introduced trees were observed and no evidence of higher male reproductive success was found for introduced trees compared with native ones either. A small but significant genetic structure was found (φft=0.05) and did not correspond to an isolation-by-distance pattern. However, we observed a significant temporal genetic structure related to the different phenological groups, especially with early and late flowering native trees; each phenological group was pollinated with distinctive pollen sources. Implications of these results are discussed in relation to the conservation and invasiveness of ash and the spread of resistance genes against pathogens such as the fungus Chalara fraxinea that is destroying common ash forests in Europe.
Similar content being viewed by others
Introduction
Afforestation and reforestation programmes are among the largest sources of genetic contamination of local populations when there are weak barriers to intra- or interspecific gene flow between introduced and local individuals (Barbour et al., 2010; Meirmans et al., 2010; DiFazio et al., 2012). In such programmes, hundreds of hectares are often planted with very few species at a high density, and thus the introduced trees are sources of a huge amount of pollen. As a consequence, in terms of the siring of seed, plantation trees if interfertile with native ones can potentially fertilize a large proportion of seeds among the surrounding native individual trees.
Intra- or interspecific gene flow (hybridization) is often seen as a source of genetic contamination (Rhymer and Simberloff, 1996) because it might lead to the introgression of non-locally adapted genetic variation and thus decrease the mean fitness of the population (Keller et al., 2000). Interspecific gene flow might also be a real threat for many native species with less rigorous reproductive barriers, especially if the resulting hybrids are more competitive and more reproductively successful than the parents (Vila et al., 2000). For conservation purposes, it is therefore important to distinguish between natural hybrids that might need to be protected in situ (Frascaria-Lacoste et al., 2011) and hybrids that have occurred because of human intervention, and which are regarded as a threat to local populations (Allendorf et al., 2001).
In certain cases, local populations that show low levels of genetic diversity because of reduced population size, inbreeding or high selective pressures, can benefit from gene flow of introduced populations (Byrne and Macdonald, 2000). Gene flow allows the spread of adaptive traits among and within populations such as those necessary to respond to pathogens. The potential of disease resistance genes to spread among ash populations is currently under intense scrutiny because of the threat posed by the ascomycete fungus Hymenoscyphus pseudoalbidus (anamorph Chalara fraxinea) that has caused the epidemic dieback disease of European ash, Fraxinus excelsior L. and F. angustifolia Vahl. (Timmermann et al., 2011; McKinnney et al., 2012).
Hybridization and invasion by hybridization will occur when features that prevent gene flow between two taxa disappear; such features can be classified as geographical or reproductive isolation factors (Arnold, 1997). Geographical isolation refers to the presence of physical barriers or geographical distance between two populations (Seehausen, 2004), whereas reproductive isolation relies on pre- or postzygotic biological mechanisms (Arnold, 1997). The introduction of non-native species is the first step in the process of invasion, and involves the transgression of natural geographical barriers between native and introduced species. The first hybridization event (production of F1 hybrids) is one of the key events required for introgression and invasion by hybridization to occur (Arnold, 1997). Therefore, it is likely that the process of introduction of hybrid individuals, in particular into new sites, will facilitate such events because there is an absence of reproductive isolation (if the hybrids are cross-fertile with the parental species) and the geographical isolation has been broken.
Evaluation of the extent of, and potential for, gene invasion in native plant populations depends largely on the ability to track the location of pollen donors. Trees are often characterized by long-distance movement of pollen, which is not necessarily homogeneous across the landscape or throughout the flowering season (Bâcles et al., 2005; Byrne et al., 2008). Pollen dispersal has been shown to be influenced by various factors, including population size and density (Willenborg et al., 2009), breeding system (Lowe and Abbott, 2004), flowering phenology (Sampson and Byrne, 2008) and variability in fecundity among individuals (Klein et al., 2008). Consequently, the amount of pollen contamination can be highly variable in space and time between local and alien populations.
Mating systems, or the proportion of selfed and outcrossed reproductive events and the mean correlation of paternity (i.e. the portion of siblings among outcrossed maternal progenies that share the same father; Ritland, 1989), have an impact on the proportion of hybrids among alien and native populations. First, introduced populations that are capable of selfing can be demographically successful by producing fertile seeds that can establish and reproduce later with local native populations. Second, populations with high rates of selfing do not show high levels of introgression, even in hybrid zones. Moreover, in wind-pollinated species, the level of outcrossing is related to the quantity of external pollen available (the mass action law, sensu Holsinger, 1991). Thus, mating systems are highly variable and depend on the density of populations (Restoux et al., 2008) and on their phenological synchrony (Fukue et al., 2007). As such, the extent of pollen dispersal can influence the mating system of trees when the rate of selfing is not controlled genetically and only depends on the relative amounts of selfed and outcrossed pollen on stigmas. This is important in hybrid zones in which there is little or no overlap in the periods of pollen emission and female receptivity between different populations, which would limit hybridization even with an apparently high density of potential parents.
One potential case of genetic invasion by alien species and hybrids has occurred in Ireland following the introduction of substantial planting grants by the Irish government in 1992 to encourage afforestation by farmers, with an extra premium for planting hardwood species such as ash. Fraxinus excelsior (common ash) is the only native ash species in Ireland. However, in some years there was a shortage of ash for planting and common ash was imported to meet farmers and foresters’ needs. After a few years, some of the planted trees were found to have poor stem form and unusual growth. In addition, the terminal buds were often pale in colour, brown rather than black and hence were more characteristic of the narrow-leaved ash, Fraxinus angustifolia. The ash plants that had been introduced into Ireland and that showed unusual morphology were identified using several molecular and morphological criteria as follows: (a) hybrid trees between F. excelsior and F. angustifolia, (b) pure F. angustifolia and (c) pure F. excelsior (Thomasset et al., 2013). Today, there are an unknown number of afforested sites in Ireland with suspected alien ash from the plantings made between 1992 and 2000. The introduced trees have started to produce flowers and seeds and in general are established close to native ash populations, raising concerns about genetic pollution via pollen- or seed-mediated gene flow.
F. excelsior and F. angustifolia have large contact zones in Europe and there is considerable potential for hybridization between the two species (Fernández-Manjarres et al., 2006; Gérard et al., 2006a; Thomasset et al., 2011b). The contact zones run from Turkey to northern Spain and along the Mediterranean basin (Thomasset et al., 2011b). Artificial control crosses (Raquin et al., 2002a) and studies on hybrid zone populations in France (Gérard et al., 2006b) have demonstrated hybridization between the two species. Both species are wind-pollinated and have a complex mating system (polygamous for F. excelsior and andromonoecious for F. angustifolia). Strictly male individuals, strictly female individuals and hermaphrodite intermediates coexist within populations of F. excelsior (Morand, 2003). In both species, the fruit consists of winged seeds (samaras) that are dispersed by wind. In continental Europe, F. angustifolia usually flowers a few months earlier than F. excelsior (Raquin et al., 2002b; Jato et al., 2004). However, an overlap in flowering times of the two species has been recorded in the Loire hybrid zone (France), but in this case assortative mating produces significant genetic isolation (Gérard et al., 2006b). Pollination can occur over short distances in dense stands in which highly fertile trees might dominate the pollen clouds (Morand, 2003) to several kilometres in small and isolated populations (Bâcles et al., 2005). A deficiency of heterozygotes, very low rates of selfing and high levels of pollen dispersal with relatively moderate degrees of isolation-by-distance are common traits that are found in different populations of F. excelsior (Morand et al., 2002; Heuertz et al., 2003; Morand, 2003). Levels of heterozygosity and gene flow as well as the mating system of F. angustifolia are much less well characterized outside hybridization studies (Fernández-Manjarres et al., 2006; Gérard et al., 2006c). However, F. angustifolia seems to show lower levels of genetic diversity within and higher geographic differentiation among isolated populations than F. excelsior (JF Fernández-Manjarrés, personal observation).
In the study reported herein, we used a combined approach that included both phenological observation and genetic analysis to determine the mating patterns and the spatial and temporal genetic structure of pollen pools of native F. excelsior and non-native ash material that had been introduced into plantations. Specific questions that we asked were: (a) Is there gene flow between introduced and native ash trees? If yes, how far with respect to plantations can pollen travel in the surrounding landscape? (b) Do introduced trees have a higher reproductive success than native ones? (c) Do introduced trees exhibit significant levels of selfing that would increase their chances of producing viable seeds? Finally, (d) do we have any evidence of spatial or temporal structure that would increase or decrease the probability of genetic invasions? On the basis of our results, we discuss the implications of the eventual contamination experienced by native Irish populations and the potential of gene flow to facilitate the transfer of adaptive traits such as disease resistance between species via hybridization.
Materials and methods
Study material and sites
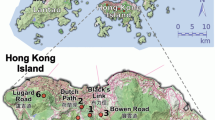
Two plantations were studied in the northwest of Dublin, Ireland, which consisted of imported trees with poor stem form, making it undesirable for commercial use. The first plantation is situated in Kildalkey (53°34′17.8″N and 6°54′54.21″W) (Figure 1a) and consists of 20 ha planted with provenances attributed mainly to northeast France (Alsace). The second plantation is located at Clonee (53°28′0309″N and 6°28′58.9″W) (Figure 1b), is more than 3.5 ha and was also planted in 1999 (imported material of unknown provenance). Trees at these sites produce flowers and seeds. Both stands are surrounded by populations of mature native common ash (F. excelsior) growing in hedgerows (Figure 1).
Maps of the two ash plantation sites in Ireland. The Clonee study site is shown on the left and Kildalkey on the right (areas in dark grey). In the case of the plantation trees, only the maternal trees sampled are represented (green triangles). All sampled native trees are represented: maternal native trees (blue squares) and other native adult trees (black circles).
Sampling
We sampled leaves from 192 adult ash trees from each plantation and 96 native ash trees around each site as potential fathers. Trees were sampled following transects that started at a random corner in each plantation, with trees that were closest to grid points spaced 30 m apart along the transects being sampled, in accordance with the sampling design described by Thomasset et al. (2013). The position of each tree was recorded using GPS coordinates and leaf samples were excised as discs (1 cm diameter) and placed in 1:1 ethanol and acetone solution for 48 h. The leaf samples were drained and then air-dried in a laminar-flow cabinet, and finally stored in microcentrifuge tubes before DNA extraction.
From among the 192 trees selected above, we chose 20 mother trees from each plantation. For each mother tree, we sampled 20 seeds from different inflorescences that resulted from open pollination; thus, a total of 400 seeds were collected per plantation (Table 1). We were only able to include a small number of mother trees because, owing to their young age, only a limited number of trees at each site were setting seed. At the Clonee site, some of the mother trees selected produced very few seeds and some of them only produced seeds that either lacked embryos or were infested by insect larvae. After DNA extraction and polymerase chain reaction (PCR) amplification, a total of 357 seeds were included in the study for the Clonee site, from the initial collection of 400 seeds.
More than half of native trees produced seeds, and therefore the sampling design was set up to include a greater number of trees with fewer seeds taken from each individual tree rather than a few large progenies, as recommended for landscape-level studies (Austerlitz and Smouse, 2002; Miyamoto et al., 2008). Thus, eight seeds were sampled per tree for 45 and 47 maternal native trees at the Clonee and Kildalkey sites, respectively (Figure 1 and Table 1).
Phenological data
A preliminary survey showed no difference between native F. excelsior flowers and flowers of the trees from plantations. All observed flowers were hermaphrodite or male type (Thomasset 2011), not related to any group in particular and with a clear absence of pure female flowers. The floral phenology of maternal trees was recorded every 10 days during the flowering season from mid-December 2009 to mid-April 2009. Floral phenology was scored using a scale from 0 to 5 (0 for the dormant flower bud stage to 5 for the seed stage/anther necrosis stage); stage 2.5 is the point of anther dehiscence/pollen release as determined by visual inspection (Supplementary Material 1). The phenology of the trees was categorized into one of three classes depending on the date at stage 2.5, just before pollen release: (1) early flowering, that is, stage 2.5 was reached before 20 March; (2) intermediate flowering (between 20 March and 20 April); and (3) late flowering (after 20 April). However, none of the sampled trees from the plantation in Kildalkey flowered late and, as consequence, plantation trees in Kildalkey were only divided into two classes (early and intermediate). Additional observations of trees in 2010 revealed that, although dates of onset vary between years, early trees are the first to flower each year, whereas late trees flower consistently at the end of the season (Thomasset, 2011).
DNA extraction and genotyping of microsatellite markers
Seeds were air-dried for 2 weeks at room temperature, rehydrated slightly overnight in water at 4 °C, immersed in 0.3 M NaOH at room temperature for 20 min and then sterilized for 2 h in calcium hypochlorite (70% active chlorine) at 4 °C (Raquin et al., 2002b). Seed wings were removed and the seeds were checked for the presence of whole embryos using a stereomicroscope. Darkened seeds were discarded because, in general, this indicated destruction by fungi or insects. Seeds were sliced into sections to improve grinding during the DNA extraction process, which might otherwise be impeded by their elasticity. To maintain uniformity in the quantity of tissue used, two-thirds of large seeds were retained, but the entirety of small seeds was used. Total DNA from 1493 seeds and 576 leaves from adult trees from both stands and surrounding areas was extracted using DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) in accordance with the manufacturer’s instructions. The quality and quantity of DNA (from 10 to 50 ng μl−1) were checked for 10% of the samples using a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, USA).
Six nuclear microsatellite markers (FEMSATL 4, FEMSATL 11, FEMSATL 12, FEMSATL 16, FEMSATL 19 (Lefort et al., 1999) and M2-30 (Brachet et al., 1999)) were used to genotype all the samples; these markers had been used previously for successful characterization of common and hybrid ash populations (Fernández-Manjarres et al., 2006; Thomasset et al., 2011a). The PCR methodology followed Morand et al. (2002). PCR products were analysed using an Applied Biosystems 3130xl Genetic Analyser (Applied Biosystems, Foster City, CA, USA) and allele sizes were determined from the raw data using GeneMapper v. 4.0 (Applied Biosystems). Individuals with doubtful and/or missing peaks were genotyped a second time. If a mismatch between the genotypes of the mother and offspring was observed, we repeated the genotyping (on mother trees and seeds) to obtain congruent maternal and offspring genotypes for all loci and avoid scoring genotypes with null alleles produced by PCR artefacts.
Data analysis
Genetic diversity
The genetic diversity of adult trees and their seeds was quantified using standard genetic statistics, including effective number of alleles (Ae), observed heterozygosity (Ho) and Hardy–Weinberg expected heterozygosity (He). These values were computed using GenAlEx 6.3 (Peakall and Smouse, 2006). The reduced sampling of native populations compared with plantation trees might bias the estimation of the mean number of alleles (allelic richness, A). We used the rarefaction method available in HP-Rare (Kalinowski, 2005) to correct for this sampling artefact. For each site progenies and adult trees were analysed at the same time; the parameter g (number of alleles sampled for diploids) was two times the minimum of individuals sampled (i.e. 192). Departures from Hardy–Weinberg equilibrium were determined using GENEPOP web 3.4 (Raymond and Rousset, 1995) using the exact probability test. Inbreeding coefficients (FIS) and their significance (1000 permutations) were computed using FSTAT 2.9.3.2 (Goudet, 2001). To control for differences in sampling scheme between progenies from planted or native trees, we repeated the genetic diversity analysis on a randomly resampled data set containing eight seeds from each of 20 mother trees per native or plantation stand. The frequencies of null alleles and the paternity exclusion probability for the second parent were estimated using CERVUS 3.03 (Marshall et al., 1998; Kalinowski et al., 2007). Moreover, we used CERVUS to estimate the error rate for incompatibility between the offspring and the known mother, which can be seen as the scoring error rate. Genetic differentiation among adult populations was evaluated with overall and pairwise FST. The significance of pairwise FST values was evaluated using 1000 permutations with the FSTAT 2.9.3.2 software.
Mating system
Mating system parameters were estimated using the maximum-likelihood procedure implemented in the MLTR 3.2 software (Ritland, 2002). The model is based on a mixed mating system in which offspring can be derived from either selfed or outcrossed fertilization (s and t). The genotypes of the maternal trees were determined based on the genotypes of the corresponding leaves. We estimated multilocus outcrossing rates (tm) and the average single-locus estimate (ts) according to groups identified according to the sampling site (Clonee or Kildalkey), whether they were plantation or native trees, and their flowering score (early, intermediate or late). The extent of biparental inbreeding was computed using the difference between single and multilocus estimates of the outcrossing rate (tm−ts) (Ritland, 2002). Using the same experimental design, we also estimated the correlation of paternity within the outcrossing pollen pools (i.e. the probability that two randomly selected offspring from the same mother shared the same father; rp(m)). This correlation was translated into an estimate of the effective number of pollen donors (Nep=1/rp(m)). Standard errors were computed using 1000 bootstrap iterations.
Paternity assignment
Paternity analysis was performed using CERVUS 3.03 (Kalinowski et al., 2007). Expanding our sampling in the surroundings will allow us to know whether gene flow between native and plantation trees may occur and at what spatial scale. However, our non-exhaustive sampling scheme will not characterize the absolute pollen contribution of each tree in populations. Seed paternity was assigned using a maximum-likelihood method with an 80% threshold (a relaxed confidence level; Marshall et al., 1998). Assigned paternity was tested if the difference between the log of odds score of the two most likely candidate fathers exceeded a certain threshold of Δ, which was computed by simulation to enable the correct assignment of a unique father in 80% of cases (Meagher, 1986). Input simulation data to determine the level of confidence in the assignments of parentage were computed using 10 000 cycles with all adult trees in the population included as candidate fathers. Although all genotypes were double-checked and some PCR reactions were repeated, we set the scoring error to 1% based on the error rate for incompatibility between the offspring and the known mother. On the basis of the missing data rate, the minimum number of loci required for assignment was set to four. Given the high density of introduced trees and the long-distance gene flow that can occur in Fraxinus (Bâcles et al., 2005), we set parameters to instruct CERVUS that about 10% of potential fathers were sampled. Pollination distances were computed as the distances between mother trees and the most likely candidate pollen donor parents. The trees in each plantation were designated as F. angustifolia, F. excelsior or hybrid groups. Designation was based on previous multivariate morphological and molecular classification (Thomasset et al., 2013). Finally, we applied a non-parametric χ2 test to compare the numbers of seeds that were sired by plantation trees classified either as F. excelsior or as hybrid type.
Spatial genetic structure of pollen pools
The spatial genetic structure of the pollen pool at the two study sites was analysed using the KinDist method (Robledo-Arnuncio et al., 2006) implemented in the software package POLDISP (Robledo-Arnuncio et al., 2007). The former method relies on the ψft, a differentiation index, derived from an analysis of molecular variance computation on the male gametes. The φft can be translated into the correlation of paternity Ψ(z) (i.e. the probability for two randomly drawn seeds to have been pollinated by the same father), and when associated with an isolation-by-distance model allows for real-time pollen dispersal estimation (Austerlitz and Smouse, 2001). We used POLDISP software to: (i) compute global and pairwise φft (Austerlitz and Smouse, 2002) for each pair of maternal sibships within each site (Clonee and Kildalkey), (ii) compute φft per phenological group and for each population: native or plantation and (iii) estimate the pollen dispersal function assuming an isolation-by-distance model. We also computed relative measures of correlated paternity between maternal sibship pairs using the similarity index for the male gametes Ψ(z) separated by a distance z. If pairwise genetic indices (Ψ) and pairwise geographical distances of all pairs of mother trees are significantly correlated, the KinDist method enables the parameters of dispersal function to be computed independently of the effective density of pollen contributors. Hence, we used a sequential method, as suggested by Robledo-Arnuncio et al. (2007), in which the pollen dispersal function was estimated first using KinDist and the effective density was estimated on the basis of the previously estimated pollen dispersal function.
Temporal structure of pollen pools
To examine the spatial and temporal structure of the pollen pools, we estimated the gametic contributions of the pollen donors directly with multivariate analysis. First, we inferred the contribution of the male gamete by subtracting the contribution of the female gamete from the genotype of the offspring. However, when mother–offspring pairs share the same heterozygote genotype for a given locus, it is not possible to determine the paternal contribution. To resolve this issue, we used a fractional probability for paternal inheritance of each of the potential alleles. For ambiguous cases, given an offspring genotype for a particular locus (A, B) and the same genotype for the mother tree (A, B), the frequencies of each allele, f(A) and f(B), were computed using the frequencies of alleles A and B in the adult population. Thus, the probability that the paternal contribution was allele A is  , whereas the probability that the paternal contribution was allele B is
, whereas the probability that the paternal contribution was allele B is  . When there was no ambiguity, the probability that an allele was inherited from the father was scored as one. A Perl script for calculating the fractional probability for paternal inheritance of each of the potential alleles for each individual is available upon request from M Thomasset. To compute the genotype frequencies, we considered all loci to be unlinked and multiplied the frequencies of each allele for all combinations of alleles over all loci. We then used the genotype frequencies of each fertilizing pollen pool to discriminate them according to their group (plantation vs native) and their flowering score (spatial structure), using a canonical discriminant analysis conducted with the R software (2009; R Foundation for Statistical Software, Vienna, Austria) and the package ade4 (Dray and Dufour, 2007).
. When there was no ambiguity, the probability that an allele was inherited from the father was scored as one. A Perl script for calculating the fractional probability for paternal inheritance of each of the potential alleles for each individual is available upon request from M Thomasset. To compute the genotype frequencies, we considered all loci to be unlinked and multiplied the frequencies of each allele for all combinations of alleles over all loci. We then used the genotype frequencies of each fertilizing pollen pool to discriminate them according to their group (plantation vs native) and their flowering score (spatial structure), using a canonical discriminant analysis conducted with the R software (2009; R Foundation for Statistical Software, Vienna, Austria) and the package ade4 (Dray and Dufour, 2007).
Results
Genetic diversity and marker resolution
All six loci analysed were highly polymorphic and a total of 196 alleles were detected (Table 1). The average allele number per locus was 22.7, ranging from 7 (FEMSATL16) to 47 (M2–30) alleles per locus (data not shown). Fifteen alleles that were unique to the Clonee plantation were identified in adult trees, with a mean of 2.5 unique alleles per locus, and eight unique alleles were identified in the Kildalkey plantation, with 1.3 unique alleles per locus on average. The frequency of unique alleles was always low and ranged from 0.003 to 0.008 (data not shown). At each site, within the adult populations (plantation and native), the values of the allelic richness were similar with approximately 21 alleles (Table 1). The mean number of alleles observed within the progenies from the native mother trees was similar to the values observed within the progenies from the plantation mother trees, and consisted of approximately 18 alleles in the offspring (Table 1). Allelic richness was a little lower within the offspring than adult trees.
All populations of adult trees showed high levels of genetic diversity (He=0.767–0.818; Table 1), and these were similar to the values found in the populations of progeny (0.777–0.805; Table 1). However, despite the high levels of diversity, all populations of adults and seeds showed significant inbreeding. Adult populations showed a positive and significant inbreeding coefficient, which indicated an excess of homozygotes (FIS from 0.053–0.147; Table 1), irrespective of their origin. The inbreeding coefficient of the progeny populations also showed a significant and positive deviation from zero (FIS from 0.104 to 0.131; Table 1). The genetic parameters (FIS, He, Ho, A, Ae) were also calculated on a data subset to take account of differences in progeny sizes (as described in the Materials and methods section). The results indicated (data not shown) that inbreeding coefficients and the levels of genetic diversity were similar to the parameters evaluated above. As genetic diversity was not affected in the subset, we kept the full data set (all progenies that we have genotyped) for the subsequent analysis. Low but significant differentiation (FST) was observed among adult populations for five of the six pairwise comparisons (Table 2); the two native Irish populations were not differentiated significantly at the P<0.01 level.
The total exclusion probabilities for paternity over the six loci were 0.997 and 0.996 for Clonee and Kildalkey, respectively. Very low levels of non-Mendelian segregation between mother and offspring genotypes were recorded and in consequence no obvious presence of null alleles was observed. However, CERVUS estimated a positive null allele frequency based on the allele frequencies of the whole data set with an average of 0.093. A low degree of variation of the null allele frequency between native and plantation trees and between sites was observed. The locus FEMSATL4 always showed the highest null allele frequency (ranging from 0.134 to 0.151) and FEMSATL19 the lowest (ranging from 0.013 to 0.018). However, CERVUS estimates null alleles frequency by analysing deviations from Hardy–Weinberg equilibrium as a consequence of high inbreeding coefficient might influence the estimation of null allele frequency. After the second genotyping, none of the mother trees for each site had missing data. For the offspring, three individuals at Clonee and Kildalkey had missing data at two loci, and 27 and 47 offspring had missing values at a single locus for Clonee and Kildalkey, respectively. After repeating the genotyping, the mismatch between the genotypes of the mother and offspring could be estimated. The results indicated that incongruence occurs on average at a frequency of approximately 0.01.
Mating system
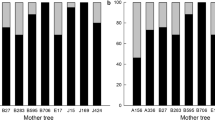
All sampled maternal trees were largely outcrossing, with similar outcrossing rates (tm) between plantation and native trees (Table 3). Moreover, comparisons of outcrossing rates among phenological groups revealed that flowering time did not affect the population-level mating system (Table 3). At the tree level, evidence of self-fertilization was found in some trees but these were not found preferentially in the native or in the plantation trees. In Clonee, only two trees from the plantation showed self-fertilization and both exhibited intermediate flowering. Three native trees (two with intermediate and one with late flowering times) showed outcrossing rates of 0.625, 0.750 and 0.875, respectively. At Kildalkey, three plantation trees showed self-fertilization; two of them flowered early (tm=0.685 or 0.95), whereas the third flowered at an intermediate date (tm=0.95). Only one native tree showed a moderate level of selfing at this site (tm=0.875).
The number of effective pollen donors calculated from the correlated paternity coefficient (rp(m)) was moderate to high at both sites, with Nep ranging from 8.6 to 75.6. It was much higher for the offspring of plantation trees than for native trees, and increased in trees that flowered later. Across all the populations analysed, only a small proportion of siblings shared the same father (rp(m) ranging from 0.015 to 0.115; Table 3). When trees were grouped according to their phenological class, offspring from early-flowering trees had a smaller number of effective pollen donors, whereas late-flowering trees showed a lower correlated paternity coefficient and a higher number of effective fathers (Table 3). Finally, a moderate level of biparental inbreeding in all populations was found regardless of the phenological class. Nevertheless, biparental inbreeding tended to be higher for the native trees than for the plantation trees (Table 3).
Gene flow
Paternity assignment
At the Clonee site, CERVUS assigned a unique father to 108 of the 717 analysed seeds (15%). At Kildalkey, a unique father was assigned to 89 of the 776 offspring (11.5%). No unique father could be assigned to 609 seeds from Clonee and 687 seeds from Kildalkey, respectively, because more than one putative father was identified or because there were no compatible male parents in the samples studied.
No evidence of an increased mating success was found for the introduced trees (χ2 test between the proportion of seeds sired by native and alien trees, regardless of mother type for both sites: χ2=0.986, d.f.=3, NS). Among seeds with an assigned unique father, offspring of native and plantation trees were sired by both native and plantation trees (Table 4). At the Clonee site, native trees sired 37/59 of offspring from plantation trees and 30/49 of offspring from native trees. At Kildalkey, 25/36 of offspring from native mothers were sired by native trees. However, for the offspring of Kildalkey plantation mother trees, the potential father was 29 times out of 53 from the plantation (Table 4).
The average pollination distance at Clonee and Kildalkey was 206 m and 285 m, respectively (Table 4). The minimum pollination distance identified between a maternal tree and an assigned pollen donor tree was 12 m, whereas the maximum distance was more than 1 km. Fewer than 35% of mating events occurred at distances of less than 100 m (Figures 2a and b). More than 30% of the pollen received by native mother trees came from trees that were located further than 400 m away (Figures 2a and b), which corresponded approximately to the size of the plantations.
For the small and dense plantation of Clonee, the frequency of pollination events observed in planted maternal trees decreased with increasing distance (Figure 2). However, for the larger and sparser plantation of Kildalkey, the peak of pollination frequency was found to be in the second distance class (100–200 m), again followed by a monotonic decrease for greater distances. With respect to native trees, no clear links between distance classes and pollination events were observed (Figure 2a). Nevertheless, at Kildalkey site where trees were more sparsely spaced, pollination events originated more frequently at greater distances than for plantation trees (Figure 2b).
We did not observe a clearly dominant paternal tree in the studied landscapes and most of the identified candidate pollen donor trees were found to have sired one or two seeds among those analysed (Table 4). For the unambiguously identified pollen donors, only one plantation tree and two native trees sired more than two seeds at Clonee, whereas one native tree sired four seeds at Kildalkey.
Gene flow between hybrid alien and native trees
Native trees were pollinated by both native and plantation trees, which included pollen donors that were assigned as hybrids (Thomasset et al., 2013). Trees identified as hybrids within the plantation sired 9 out of 19 offspring (47.4%) collected from native mother trees at Clonee. In Kildalkey, 7 out of 11 (63.6%) offspring collected from native mothers had a pollen donor identified as a hybrid. There were no significant differences between the number of native mother trees pollinated by hybrid trees and those pollinated by trees characterized as F. excelsior inside the plantations (χ2=0.42, d.f.=1, P=0.515 for Clonee; χ2=3.53, d.f.=1, P=0.060 for Kildalkey).
Spatial structure of the pollen pools
Global φft among pollen pools sampled by female trees were low and similar for the two sites (φft=0.055 and φft=0.050 at Kildalkey and Clonee, respectively). This indicated that more than 94% of the genetic variability of the fertilizing pollen pool was found within the offspring from any given mother. Pairwise φft values were also low on average, although high values of φft were observed for certain trees, mostly among native individuals (Figure 3). Indeed, pairwise φft values were slightly higher within the native tree groups (0.062±0.049 and 0.066±0.045 for Clonee and Kildalkey, respectively) than within the plantation tree group at both sites (0.037±0.027 and 0.040±0.035 for Clonee and Kildalkey, respectively). Pairwise φft values among native and plantation trees were intermediate (0.051±0.036 and 0.056±0.043 for Clonee and Kildalkey, respectively; data not shown).
Similar results were observed for Ψ. The KinDist method indicated a low but significant genetic structure. However, neither site showed an isolation-by-distance structure for pollen because the Spearman’s rank correlations for correlated paternity among sibships (Ψ(z)) and the distance z between two mother trees were negative, at −0.05 and −0.04, respectively, and were not significantly different from 0. Hence, it was not possible to compute the pollen dispersal function with POLDISP. Similar results were observed when data were partitioned into three phenological groups (early, intermediate and late flowering), which indicated again that, even in each phenological class, there was no spatial decline in the relatedness of the pollen donors with increasing distance.
Temporal structure of the pollen pools
At both sites, the composition of the pollen pools was different between the plantation and native populations based on the canonical discriminant analysis of fertilizing gametes. Pollen cloud received by native trees from the three phenological classes tended not to cluster with the pollen received by the plantation trees. Moreover, the same analysis showed that pollen pools could be differentiated partially according to the three phenological groups (Figure 4). Even if native and plantation trees with intermediate phenology shared their pollen pools (Figures 4a and b), native mother trees from early and late phenological groups clearly sampled different pollen pools at both study sites. However, early-flowering plantation trees in Clonee and Kildalkey shared part of their pollen pool with the intermediate-flowering trees.
Plot of the two first canonical scores of the canonical discriminant analysis (CDA) at Clonee (a) and Kildakey (b) sites on the pollen gametes of Fraxinus spp. in Ireland. The pollen cloud differentiation for each mother tree is shown. Trees were grouped according to their flowering phenology (Int for intermediate dates of flowering; Early and Late for the two other flowering groups) and their sampling location (plantation trees in different shades of grey and native trees in different shades of blue). The figure shows 95% confidence intervals around the mean canonical score for the different groups.
Discussion
The results of our study showed that native mother trees surrounding plantation sites were pollinated by either individuals from the plantations or by other native trees, the extent of the pollination being limited by the phenology of the trees. Moreover, all types of trees in the plantations, F. angustifolia, F. excelsior, and ash of hybrid origin, could pollinate native trees but there was no evidence of reproductive advantage of introduced trees. Genotyping all the fathers present in the area (in the surrounding landscape and plantation trees) will probably decrease the number of unidentified fathers and give a more precise picture of pollen exchange between plantations (hybrids and non-hybrid types) and the native trees. At the time of the study, the plantation trees were just reaching the adult flowering stage of their life cycle and were much younger in age than the native trees (they produced far fewer flowers; Thomasset, 2011). Furthermore, approximately half of the plantation trees had not yet started to flower (Thomasset, 2011). In the near future, all the plantation trees at these sites should be able to reproduce. On the basis of our results, pollen from plantation trees would be expected to compete strongly with pollen from native trees.
How far does pollen disperse?
Our two study sites were located in windy open farmland landscapes within fragmented rather than continuous forest, suggesting few limits to gene flow. We found that most matches between unambiguously identified pollen donors and offspring occurred at distances between 100 and 299 m in the case of plantation trees (Figures 2a and b) and at distances of more than 200 m in the case of native trees, with a peak at distances >400 m. Moreover, mother trees were not pollinated more frequently by neighbouring pollen donors than by any random tree in the population (Figure 3). Average distances for pollen dispersal of 260 m and 382 m (between an identified pollen donor and mother tree) were found for the two native populations at Clonee and Kildalkey, respectively. These distances are similar to those found by Bâcles et al. (2005), who recorded an effective dispersal distance of 328 m within Scottish remnant ash populations. However, because of the large size of the plantation, our sampling was not exhaustive inside the sampling area and represented approximately 10% of the adult trees present. As such, our estimate of the pollen-dispersal based on paternity assignment analysis into this landscape is very much an average and might be overestimated if unsampled trees from the surrounding landscape and from the plantation contributed largely to pollination.
The occurrence of mating at the shortest distance for the plantation trees may be associated with a particular spatial configuration of trees. The higher density of trees at Clonee led to a greater number of potential fathers at a short distance, whereas the number of potential fathers located within 100 m was lower at Kildalkey, probably because the density of potential fathers was also lower in this sparse landscape (Figure 2). Contrary to what was observed at the local scale by Gérard et al. (2006c) and Bâcles and Ennos (2008) for F. excelsior, our probabilities of pollen dispersal did not decrease rapidly with increasing distance from the pollen source for native trees at both sites. Our limited ability to estimate the parameters of the dispersal curves may be due to our sampling scheme, or to high rates of pollen immigration from trees outside the study site. Indeed, the two earlier studies mentioned above also showed the presence of high levels of immigrant pollen (i.e. from outside the sampled area), and long-distance pollen dispersal following a fat-tailed dispersal kernel. In the present study, the spatial scales at which the decline in relatedness will appear remain unknown. Finally, the spatial configuration of trees at Kildalkey, where the plantation is large and native trees are located at an average distance of approximately 400 m from other trees (see Supplementary Material 2), may explain the peak of pollination events at this distance for native trees (Figure 2b).
For species that show a wide-ranging dispersal of pollen, such as ashes, it is more likely that the spatial distribution of genotypes will be close to random than for those with limited dispersal (Chung et al., 2000). Indeed, in anemophilous species, pollen can move further in landscapes that are subject to deforestation and fragmentation than in landscapes with continuous forest (Bâcles et al., 2005; Sampson and Byrne, 2008; Nielsen and Kjaer, 2010). Hence, introduced plantation trees could represent a clear source of risk for this type of open mixed agricultural and forest landscape in Ireland.
However, alien gene flow could also prove as a source of adaptive traits for abiotic or biotic stresses such as those likely to occur with climate change (Thomasset et al., 2011b) or those currently being generated by the epidemic ash dieback disease C. fraxinea (Kowalski 2006; Kirisits et al., 2010). Problematic ash dieback was first recorded in Poland in 1992 and has since spread over large geographic distances. By the end of 2010, it had been recorded in over 20 European countries (Timmermann et al., 2011). As ash trees are long lived and typically take approximately 10 years to flower, the spread of ash resistance at a population genetic level cannot match the speed of pathogen spread. Disease resistance has been recorded in ash (McKinney et al., 2012) and the speed of population recovery will largely depend on the extent of pollen- and seed-mediated gene flow.
Pollen clouds, phenology and genetic risks
The estimated level of differentiation of the pollen cloud among mothers was low but significant (φft close to 0.05 at both sites) and consistent with previous studies on F. excelsior (φft=0.06, Morand, 2003), but lower than in hybrid zones (φft=0.11; Gérard et al., 2006c). The low pollen pool differentiation within plantation tree populations indicated that plantation trees probably have more phenological overlap and likely mated more among themselves than with native trees. Moreover, we observed a tendency for a higher level of differentiation of the pollen pools within native trees at both sites than for differentiation between native and plantation pollen pools.
Despite the lack of spatial genetic structure in the pollen cloud (Figure 3), we found evidence of partial temporal isolation between plantation and native trees. Indeed, according to our results, the structure of pollen clouds was better explained by flowering time (canonical discriminant analysis; Figure 4) and by the source of the material (native/plantation) than by spatial distance (Figure 4). The genetic structure statistics estimated from the seeds from different types of mothers showed that a significant percentage (3% for Clonee and 4% for Kildalkey, analysis of molecular variance P<0.001; results not shown) of the respective total genetic variation was explained by differences among the phenological classes of the mother trees. In fact, studies on other forest tree species (Byrne et al., 2008; Sampson and Byrne, 2008; Field et al., 2011) have highlighted the fact that variation in the source pollen pool can be attributed to flowering phenology and not just to geographical distances between populations. Gérard et al. (2006c) found that earlier-flowering male ash trees participated to a greater extent as pollen donors and had a higher degree of mating success than later-flowering individuals in a natural hybrid zone. Moreover, for mature trees in the same study, genetic differences increased with increasing difference in flowering phenology.
In the present study, we observed that the number of effective fathers was always higher for seeds from mother trees that flowered late (Table 3). The observed pattern of an increasing number of effective fathers as the flowering season advanced can be explained by the increase in availability of mates. Surprisingly, results for Clonee showed that late-flowering native F. excelsior individuals (Figure 4a), which can potentially receive pollen from trees that flowered earlier, had a distinct pollen cloud. The fact that the number of effective fathers was higher in this phenological group than in the trees with an earlier flowering period (Table 3) could confirm the occurrence of long-distance pollen movements. However, for Kildalkey that is sparse and for which we would expect many long-distance pollen events, the pollen pool of native late-flowering mother trees overlapped with that of native trees with an intermediate flowering time (Figure 4b). Whether the high number of pollen donors is related to the total size of the local populations or to long-distance pollen flow at the peak of the flowering season for F. excelsior cannot be evaluated with our data. Study on two Eucalyptus species (Field et al., 2011) showed that pollen dispersal and hybridization frequencies were influenced strongly by demographic factors and flowering synchrony, which seems to be the case in Ireland also.
Finally, it is worth noting that the pollen pool of seeds from native early-flowering mother trees did not cluster with the pollen pool of seeds from early-flowering plantation trees at either sites (Figures 4a and b), despite the overlap in their flowering times. We determined phenological classes on the basis of male function and did not measure the receptivity of the stigmas. Future studies should focus on the observation and quantification of the start of female function, which might be more indicative of phenological isolation. Moreover, gametophytic or sporophytic incompatibility (depending on the factors that determine the pollen phenotype are expressed after or before the advent of the meiosis) might explain the lack of clustering of the pollen clouds of early-flowering plantation and native trees.
Mating system and invasiveness
In the present study, invasion or naturalization potential will not depend on the rate of selfing because we observed very low rates of selfing at the global level in the populations (between 1 and 2%; Table 3), which was consistent with previous studies on ash (Heuertz et al., 2003; Bâcles et al., 2005). Our results contrast with early flowering ash in natural hybrid zones that were shown to have a higher rate of selfing due to a lack of available pollen (Gérard et al., 2006c). Several studies have shown that plant species with a mixed mating system have a larger invasion range than strict outcrossers when few available mates are present in the environment (van Kleunen et al., 2008; Sloop et al., 2009). In the present case, the large number of potential mates near to and within plantations, which originated from an artificial mixture of hybrids, F. excelsior and F. angustifolia, might have provided sufficient pollen donors throughout the pollination periods.
Seeds that resulted from selfing might also be difficult to detect because they might be of suboptimal quality and might have been discarded from the genetic screening. Indeed, seeds from many of the plantation trees were infested with insect larvae and were not viable (Thomasset, 2011). Selection against selfed seeds would also argue against invasion. We cannot however use our data to assess whether seeds that are more susceptible to insect infestation tend to be more inbred.
Conclusions
Our results have shown that pollen dispersal has occurred over large distances between native and non-native Fraxinus trees both inside and outside plantations in Ireland. Consequently, genes from alien ash trees can potentially spread to native Irish ash populations not only by direct dispersal of pollen from the plantation sites but also by subsequent seed dispersal from these sites. Nevertheless, no significant reproductive advantage was observed for the introduced trees compared with the native ones. Likewise, introduced populations seem to have a low selfing rates (and do not have a greater tendency for selfing than native trees), which suggests that selfing should not have a major role in the invasion potential of the introduced continental ashes.
The results also showed that determination of the mating system or paternity analyses alone are insufficient to describe the complete pattern of interspecific gene flow. Indeed, the ability to detect temporal structure in the pollen cloud enabled us to identify differentiation within the pollen pool, which would otherwise have been overlooked. We observed structure in the pollen cloud in relation to different flowering times that could reduce, but not completely, prevent gene flow between plantation and native trees. From a conservation perspective, we advise seed managers to harvest seeds from early- or late-flowering native trees because they have been shown to receive clearly distinctive (native) pollen clouds. In addition, seed collections should be made as far away as possible from any suspect site with introduced ashes. Furthermore, foresters should avoid creating large plantations of imported material close to native wild populations whenever possible.
It remains unclear whether hybrid progeny that are produced around or within plantations will be sufficiently fit to compete in the Irish environment and to invade the landscape. Our initial finding in this regard was a high rate of predation by insects among seeds from plantation trees that might indicate non-adaptation. So far there has been no evident spread of hybrid-like seedlings in either of the plantations (M Thomasset, personal observation). Current data are insufficient to predict whether zygotes of interspecific and introgressed crosses will survive in the future. In the long term, it is also not known whether introgression will have a positive or negative effect on Irish ash populations. Introgression from non-native sources could in fact provide a novel source of genes for selection. However, simulation analyses suggest that the future Irish climate will not match the present-day climate of continental hybrid zones, casting doubts on any selective advantage of hybrids under expected future climates in Ireland (Thomasset et al., 2011b). Interspecific hybridization and natural introgression provide a route for adaptive genes such as those to pests and diseases to spread through populations. Gene flow will be critical for the recovery of ash populations to the epidemic ash dieback disease. F. excelsior and F. angustifolia are both susceptible to C. fraxinea but resistance has been found (McKinney et al., 2012). Leaf phenology, especially leaf shed and growth period, has been shown to influence susceptibility to the disease (McKinney et al., 2011). Flower phenology is related to leaf phenology in ashes (early flowering trees flush leaves first; C Raquin, personal communication). Thus, variation in flower phenology recorded here among hybrid and native ashes could be significant for natural or artificial selection of resistant genotypes.
Data archiving
Data available from the Dryad Digital Repository: doi:10.5061/dryad.55c2f.
References
Allendorf FW, Leary RF, Spruell P, Wenburg JK . (2001). The problems with hybrids: setting conservation guidelines. Trends Ecol Evol 16: 613–622.
Arnold ML . (1997) Natural Hybridization and Evolution. Oxford University Press: Oxford, UK.
Austerlitz F, Smouse PE . (2001). Two-generation analysis of pollen flow across a landscape. II. Relation between Phi(ft), pollen dispersal and interfemale distance. Genetics 157: 851–857.
Austerlitz F, Smouse PE . (2002). Two-generation analysis of pollen flow across a landscape. IV. Estimating the dispersal parameter. Genetics 161: 355–363.
Bâcles CFE, Burczyk J, Lowe AJ, Ennos RA . (2005). Historical and contemporary mating patterns in remnant populations of the forest tree Fraxinus excelsior L. Evolution 59: 979–990.
Bâcles CFE, Ennos RA . (2008). Paternity analysis of pollen-mediated gene flow for Fraxinus excelsior L. in a chronically fragmented landscape. Heredity 101: 368–380.
Barbour RC, Wise SL, McKinnon GE, Vaillancourt RE, Williamson GJ, Potts BM . (2010). The potential for gene flow from exotic eucalypt plantations into Australia’s rare native eucalypts. Forest Ecol Manage 260: 2079–2087.
Brachet S, Jubier MF, Richard M, Jung-Muller B, Frascaria-Lacoste N . (1999). Rapid identification of microsatellite loci using 5′ anchored PCR in the common ash Fraxinus excelsior. Mol Biol 8: 157–169.
Byrne M, Elliott C, Yates C, Coates D . (2008). Maintenance of high pollen dispersal in Eucalyptus wandoo, a dominant tree of the fragmented agricultural region in Western Australia. Conserv Genet 9: 97–105.
Byrne M, Macdonald B . (2000). Phylogeography and conservation of three oil mallee taxa, Eucalyptus kochii ssp. kochii, ssp. plenissima and E. horistes. Austr J Bot 48: 305–312.
Chung MG, Chung MY, Oh GS, Epperson BK . (2000). Spatial genetic structure in a Neolitsea sericea population (Lauraceae). Heredity 85: 490–497.
DiFazio SP, Leonardi S, Slavov GT, Garman SL, Adams WT, Strauss SH . (2012). Gene flow and simulation of transgene dispersal from hybrid poplar plantations. N Phytol 193: 903–915.
Dray S, Dufour AB . (2007). The ade4 package: implementing the duality diagram for ecologists. J Statist Softw 22: 1–20.
Fernández-Manjarrés JF, Gerard PR, Dufour J, Raquin C, Frascaria-Lacoste N . (2006). Differential patterns of morphological and molecular hybridization between Fraxinus excelsior L. and Fraxinus angustifolia Vahl (Oleaceae) in eastern and western France. Mol Ecol 15: 3245–3257.
Field DL, Ayre DJ, Whelan RJ, Young AG . (2011). The importance of pre-mating barriers and the local demographic context for contemporary mating patterns in hybrid zones of Eucalyptus aggregata and Eucalyptus rubida. Mol Ecol 20: 2367–2379.
Frascaria-Lacoste N, Henry A, Gérard PR, Bertolino P, Collin E, Fernández-Manjarrés JF . (2011). Should forest restoration with natural hybrids be allowed? Restor Ecol 19: 701–704.
Fukue Y, Kado T, Lee S, Ng KKS, Muhammad N, Tsumura Y et al. (2007). Effects of flowering tree density on the mating system and gene flow in Shorea leprosula; (Dipterocarpaceae) in Peninsular Malaysia. J Plant Res 120: 413–420.
Gérard PR, Fernández-Manjarrés JF, Bezrtolino P, Dufour J, Raquin C, Frascaria-Lacoste N . (2006a). New insights in therecognition of the European ash species Fraxinus excelsior L. and Fraxinus angustifolia Vahl as useful tools for forest management. Ann Forest Sci 63: 733–739.
Gérard PR, Fernández-Manjarrés JF, Frascaria-Lacoste N . (2006b). Temporal cline in a hybrid zone population between Fraxinus excelsior L. and Fraxinus angustifolia Vahl. Mol Ecol 15: 3655–3667.
Gérard PR, Klein EK, Austerlitz F, Fernández-Manjarrés JF, Frascaria-Lacoste N . (2006c). Assortative mating and differential male mating success in an ash hybrid zone population. BioMed Central Evol Biol 6: 96–110.
Goudet J . (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available at http://www2.unil.ch/popgen/softwares/fstat.htm (updated from Goudet, 1995).
Heuertz M, Vekemans X, Hausman J-F, Palada M, Hardy OJ . (2003). Estimating seed vs pollen dispersal from spatial genetic structure in the common ash. Mol Ecol 12: 2483–2495.
Holsinger KE . (1991). Mass-action models of plant mating systems: the evolutionary stability of mixed mating systems. Am Naturalist 138: 606–622.
Jato V, Rodriguez-Rajo FJ, Dacosta N, Aira MJ . (2004). Heat and chill requirements of Fraxinus flowering in Galicia (NW Spain). Grana 43: 217–223.
Kalinowski ST . (2005). HP-Rare: a computer program for performing rarefaction on measures of allelic diversity. Mol Ecol Notes 5: 187–189.
Kalinowski ST, Taper ML, Marshall TC . (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16: 1099–1106.
Keller M, Kollmann J, Edwards P . (2000). Genetic introgression from distant provenances reduces fitness in local weed populations. J Appl Ecol 37: 647–659.
Kirisitsa T, Matlakovaa M, Mottinger-Kroupaa S, Halmschlagera E, Lakatosb F . (2010). Chalara fraxinea associated with dieback of narrow-leafed ash (Fraxinus angustifolia). Plant Pathol 59: 411.
Klein EK, Desassis N, Oddou-Muratorio S . (2008). Pollen flow in the wildservice tree, Sorbus torminalis (L.) Crantz. IV. Whole interindividual variance of male fecundity estimated jointly with the dispersal kernel. Mol Ecol 17: 3323–3336.
Kowalski T . (2006). Chalara fraxinea sp. nov. associated with dieback of ash (Fraxinus excelsior) in Poland. Forest Pathol 36: 264–270.
Lefort F, Brachet S, Frascaria-Lacoste N, Edwards KJ, Douglas GC . (1999). Identification and characterization of microsatellite loci in ash (Fraxinus excelsior L.) and their conservation in the olive family (Oleaceae). Mol Ecol 8: 1088–1090.
Lowe AJ, Abbott RJ . (2004). Reproductive isolation of a new hybrid species, Senecio eboracensis Abbott & Lowe (Asteraceae). Heredity 92: 386–395.
Marshall TC, Slate J, Kruuk LEB, Pemberton JM . (1998). Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7: 639–655.
McKinney LV, Nielsen LR, Hansen JK, Kjær ED . (2011). Presence of natural genetic resistance in Fraxinus excelsior (Oleaceae) to Chalara fraxinea (Ascomycota): an emerging infectious disease. Heredity 106: 788–797.
McKinney LV, Thomsen IM, Kjær ED, Nielsen LR . (2012). Genetic resistance to Hymenoscyphus pseudoalbidus limits fungal growth and symptom occurrence in Fraxinus excelsior. Forest Pathol 42: 69–74.
Meagher TR . (1986). Analysis of paternity within a natural population of Chamaelirium luteum. 1. Identification of most-likely male parents. Am Naturalist 128: 199.
Meirmans PG, Lamothe M, Gros-Louis MC, Khasa D, Périnet P, Bousquet J et al. (2010). Complex patterns of hybridization between exotic and native North American poplar species. Am J Bot 97: 1688–1697.
Miyamoto N, Fernández-Manjarrés JF, Morand-Prieur ME, Bertolino P, Frascaria-Lacoste N . (2008). What sampling is needed for reliable estimations of genetic diversity in Fraxinus excelsior L. (Oleaceae)? Ann Forest Sci 65.
Morand M-E . (2003) Evolution et maintien d'un système de reproduction polymorphe. Approche génétique et écologique de la polygamie chez le frêne commun, Fraxinus excelsior L. PhD thesis, ENGREF, Nancy, France.
Morand M-E, Brachet S, Rossignol P, Dufour J, Frascaria-Lacoste N . (2002). A generalized heterozygote deficiency assessed with microsatellites in French common ash populations. Mol Ecol 11: 377–385.
Nielsen LR, Kjaer ED . (2010). Geneflow and matting patterns in individuals of wych elm (Ulmus glabra) in forest and open land after the influence of Dutch elm disease. Conserv Genet 11: 257–268.
Peakall R, Smouse PE . (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295.
Raquin C, Brachet S, Jeandroz S, Vedel F, Frascaria-Lacoste N . (2002a). Combined analyses of microsatellite and RAPD markers demonstrate possible hybridisation between Fraxinus excelsior L. and Fraxinus angustifolia Vahl. Forest Genet 9: 111–114.
Raquin C, Jung-Muller B, Dufour J, Frascaria-Lacoste N . (2002b). Rapid seedling obtaining from European ash species Fraxinus excelsior L. and Fraxinus angustifolia Vahl. Ann Forest Sci 59: 219–224.
Raymond M, Rousset F . (1995). (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249.
Restoux G, Silva DE, Sagnard F, Torre F, Kléin E, Fady B . (2008). Life at themargin: the mating system of Mediterranean conifers. Web Ecol 8: 94–102.
Rhymer JM, Simberloff D . (1996). Extinction by hybridization and introgression. Annu Rev Ecol Syst 27: 83–109.
Ritland K . (2002). Extensions of models for the estimation of mating systems using an independent loci. Heredity 88: 221–228.
Ritland K . (1989). Correlated matings in the partial selfer, Mimulus guttatus. Evolution 43: 848–859.
Robledo-Arnuncio JJ, Austerlitz F, Smouse PE . (2006). A new method of estimating the pollen dispersal curve independently of effective density. Genetics 173: 1033–1045.
Robledo-Arnuncio JJ, Austerlitz F, Smouse PE . (2007). POLDISP: a software package for indirect estimation of contemporary pollen dispersal. Mol Ecol Notes 7: 763–766.
Sampson JF, Byrne M . (2008). Outcrossing between an agroforestry plantation and remnant native populations of Eucalyptus loxophleba. Mol Ecol 17: 2769–2781.
Seehausen O . (2004). Hybridization and adaptive radiation. Trends Ecol Evol 19: 198–207.
Sloop CM, Ayres DR, Strong DR . (2009). The rapid evolution of self-fertility in Spartina hybrids (Spartina alterniflora × foliosa) invading San Francisco Bay, CA. Biol Invas 11: 1131–1144.
Thomasset M . (2011) Introduced Hybrid Ash: Fraxinus excelsior × F. angustifolia in Ireland and its Potential for Interbreeding with Native Ash. Trinity College: Dublin, UK.
Thomasset M, Fernández-Manjarres JF, Douglas GC, Frascaria-Lacoste N, Raquin C, Hodkinson TR . (2011a). Molecular and morphologicalcharacterization of reciprocal F1 hybrid ash (Fraxinus excelsior × F. angustifolia, Oleaceae) and parental species reveals asymmetric character inheritance. Int J Plant Sci 172: 423–433.
Thomasset M, Ferná;ndez-Manjarres JF, Douglas GC, Frascaria-Lacoste N, Hodkinson TR . (2011b). Hybridisation, introgression and climate change: a case study for the tree genus Fraxinus (Oleaceae). In: Hodkinson TR, Jones MB, Waldren S, Parnell JAN eds. Climate Change, Ecology and Systematics. Cambridge University Press: Cambridge, UK.
Thomasset M, Fernández-Manjarrés JF, Douglas GC, Bertolino P, Frascaria-Lacoste N, Hodkinson TR . (2013). Assignment testing reveals multipleintroduced source populations including potential ash hybrids (Fraxinus excelsior × F. angustifolia) in Ireland. Eur J Forest Res 132: 195–209.
Timmermann V, Børja I, Hietala AM, Kirisits T, Solheim H . (2011). Ash dieback: pathogen spread and diurnal patterns of ascospore dispersal, with special emphasis on Norway. OEPP/EPPO Bull 41: 14–20.
van Kleunen M, Manning JC, Pasqualetto V, Johnson SD . (2008). Phylogenetically independent associations between autonomous self-fertilization and plant invasiveness. Am Naturalist 171: 195–201.
Vila M, Weber E, Antonio CMD . (2000). Conservation implications of invasion by plant hybridization. Biol Invas 2: 207–217.
Willenborg C, Brûlé-Babel A, Van Acker R . (2009). Low crop plant population densities promote pollen-mediated gene flow in spring wheat (Triticum aestivum L.). Transg Res 18: 841–854.
Acknowledgements
We thank Mr Gerald Potterton and Mr Brian White for granting access to their land at Kildalkey and Clonee, respectively. We also thank Paola Bertolino, John McNamara, Jeanne O’ Callaghan, Paola Ugoletti and Frederic Coutou for their technical assistance with the laboratory work and data collection in the field. We are also very grateful to François Thomasset for his help with the Perl programming. This work is part of the PhD research of the first author, which is entitled: ‘Identifying the scale of suspected hybrid ash (F. excelsior × F. angustifolia) in Ireland and its potential for genetic pollution of indigenous ash germplasm, ASHGEN’. It was funded partially by COFORD (Council for Forest Research and Development) and by the Teagasc Walsh Fellowship Scheme.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Thomasset, M., Hodkinson, T., Restoux, G. et al. Thank you for not flowering: conservation genetics and gene flow analysis of native and non-native populations of Fraxinus (Oleaceae) in Ireland. Heredity 112, 596–606 (2014). https://doi.org/10.1038/hdy.2013.141
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2013.141
Keywords
This article is cited by
-
Pollination success of Fraxinus excelsior L. in the context of ash dieback
Annals of Forest Science (2023)
-
Characterising the molecular diversity of ash (Fraxinus excelsior L.) at its western marginal range in Europe — phylogeographic insights and implications for conservation in Ireland
Tree Genetics & Genomes (2022)
-
Gene flow and reproductive success in ash (Fraxinus excelsior L.) in the face of ash dieback: restoration and conservation
Annals of Forest Science (2021)
-
Lack of genetic structure and evidence for long-distance dispersal in ash (Fraxinus excelsior) populations under threat from an emergent fungal pathogen: implications for restorative planting
Tree Genetics & Genomes (2015)