Abstract
Game species are often manipulated by human beings, whose activities can deeply affect their genetic make-up and population structure. We focused on a geographically isolated wild boar population (Sardinia, Italy), which is classified, together with the Corsican population, as a separate subspecies (Sus scrofa meridionalis). Two hundred and ten wild boars collected across Sardinia were analysed with a set of 10 microsatellites and compared with 296 reference genotypes from continental wild populations and to a sample of domestic pigs. The Sardinian population showed remarkable diversity and a high proportion of private alleles, and strongly deviated from the equilibrium. A Bayesian cluster analysis of only the Sardinian sample revealed a partition into five subpopulations. However, two different Bayesian approaches to the assignment of individuals, accounting for different possible source populations, produced consistent results and proved the admixed nature of the Sardinian population. Indeed, introgressive hybridization with boars from multiple sources (Italian peninsula, central Europe, domestic stocks) was detected, although poor evidence of crossbreeding with free-ranging domestic pigs was unexpectedly found. After excluding individuals who carried exotic genes, the population re-entered Hardy–Weinberg proportions and a clear population structure with three subpopulations emerged. Therefore, the inclusion of introgressed animals in the Bayesian analysis implied an overestimation of the number of clusters. Nonetheless, two of them were consistent between analyses and corresponded to highly pure stocks, located, respectively, in north-west and south-west Sardinia. This work shows the critical importance of including adequate reference samples when studying the genetic structure of managed wild populations.
Similar content being viewed by others
Introduction
Genetic diversity within managed populations is affected by human beings in a number of ways. Harvest can cause the loss of genetic variation, an alteration of genetic subdivision, and selective changes with effects on the phenotype (Allendorf et al., 2008). The introduction of exotic fauna accounts for allopatric taxa, species as well as subspecies, occurring in sympatry, and this creates the conditions for introgressive hybridization (Simberloff, 1996; McDevitt et al., 2009). Translocation of individuals, also for conservation purposes, can severely alter the genetic structure of indigenous populations (DeYoung et al., 2003; Frantz et al., 2006; Latch et al., 2006), and encourage the deliberate hybridization between domestic and wild organisms (Allendorf et al., 2001). As a consequence, geographical patterns of genetic variation in a managed species should be evaluated with caution, as both genetic diversity and structure in a population might be affected by artificial modifications.
In this work, we study the effects of genetic admixture in a wild population by evaluating its genetic subdivision twice. First we consider the population itself, disregarding any possible source of immigration/introgression. Second, we consider the study population together with other reference populations, controlling for the effect of immigration/introgression.
The wild boar (Sus scrofa) is a game species that is highly managed throughout its distribution range. It almost disappeared from several regions of Europe at the beginning of the last century, subsequently recovered almost everywhere and now can be considered a pest in many countries (Apollonio et al., 2010). Despite the natural recovery capability of this species, exploited wild boar populations are often restocked with individuals from other populations with whom native boars can hybridize. Furthermore, wild populations can crossbreed with domestic pigs where open-air pig farming is practised. As a result, many wild boar populations have likely been perturbed by human beings and the interpretation of their present genetic make-up carries the risk of being puzzling.
According to zoo-archaeological records (Wilkens, 2003; Albarella et al., 2006), pig presence in Sardinia dates back to the time of the first human settlements during the early Neolithic. The wild population is supposed to have originated when Neolithic pigs escaped from man's control and became feral. The evolution in isolation led them to diverge from continental populations, so that towards the end of the sixteenth century the Sardinian wild pig was classified as a distinct subspecies (Sus scrofa meridionalis; Major, 1883), on the basis of morphological traits (body size, skull length, lacrimal bone). This classification was confirmed by subsequent taxonomic reassessments (Boetticher, 1941; Groves, 1981; Apollonio et al., 1988; Oliver et al., 1993). Crossbreeding between human-reared and wild pigs was possible for a long time in Sardinia. As a consequence, the two populations presumably shared a common gene pool. Since the sixteenth century, the diffusion of pig breeding in captivity has limited the possibility of crossbreeding, with the exception of some inner areas where extensive outdoor farming is still practised. At the same time, the traditional local pig breed has been progressively introgressed by crossings with commercial breeds that have been introduced to improve meat production (Carta et al., 2007). Likewise, anecdotal information reported the (illegal) introduction of wild boars from the mainland, especially after the Second World War. This practice, promoted by hunters and involving local breeders, represented a source of further genetic admixture, one that likely jeopardized the genetic integrity of the Sardinian subspecies. Therefore, the present-day composition of the Sardinian wild boar population is assumed to be the result of a multi-millennial history of independent evolution (geographic isolation), followed by introgressive hybridization with ameliorated pig breeds and genetic admixture with continental wild boars.
The analysis of nuclear co-dominant markers (for example, microsatellites) enhanced by improved statistical approaches allows for the assignment of an individual to the population where its genotype is most likely to have arisen (assignment tests), and to detect individuals with admixed ancestry (Vernesi et al., 2003; Choisy et al., 2004; Spencer and Hampton, 2005; Frantz et al., 2006; Lecis et al., 2006). Bayesian methods are commonly employed to infer the genetic structure of a population, either disregarding or addressing a priori assumptions about partitioning within the population, which in turn can either take into account or neglect spatial data (Pritchard et al., 2000; Corander et al., 2003; Guillot et al., 2005).
In a previous work on genetic diversity in European wild boar populations (Scandura et al., 2008), we had included a sample of wild boars and domestic pigs from Sardinia. They showed dissimilar gene pools, and while Sardinian domestic pigs clustered with samples of other pig breeds, wild boars were strongly differentiated from any continental population. The same study also showed that microsatellite genotyping makes it possible to trace the origin of single specimens, thus distinguishing among different populations/areas in Europe.
The goal of this study was to show that inferences on population structure could be misleading if the effects of human-mediated immigration/introgression are not taken into account. With this purpose, we tested the hypotheses that the Sardinian wild boar population represents an admixture of different gene pools, owing to introgression from continental wild boars and crossbreeding with domestic pigs, and that this perturbation has the potential to distort the signal of genetic subdivision.
With the aim of disentangling the misleading effect of genetic introgression on the inference of population subdivision, we performed the following steps: (1) we explored the overall present-day genetic composition of the Sardinian population according to microsatellite data; (2) we assessed the genetic nature of each individual by comparison with other European populations and with domestic pigs; (3) we excluded from the dataset all individuals suspected to be immigrants or to have admixed ancestry; and (4) we analysed the structure of the ‘purged’ population.
Materials and methods
Sampling and microsatellite genotyping
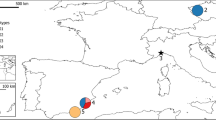
A total of 210 wild boar samples from all over Sardinia (Supplementary Figure S1) were provided by local hunters between 2001 and 2008, and stored either in absolute ethanol or frozen until analysis. DNA was isolated from tissue samples using either the QIAamp (Qiagen, Hilden, Germany) or the Genelute (Sigma-Aldrich, St Louis, MO, USA) kit. Sampling locations were plotted using ArcView GIS 3.2 (ESRI, Redlands, CA, USA), with each sample referring to the municipality where the animal was killed. Sardinian wild boars were compared with 232 wild boars from several European countries (Italy n=124, Spain n=15, France n=32, Luxembourg n=10, Austria n=13, Hungary n=20, Poland n=18). In addition, 64 domestic pigs were sampled, including 22 pigs from the historical Italian breed Cinta Senese and 37 outdoor-bred Sardinian pigs.
Ten microsatellites (S026, S215, S355, SW72, SW461, SW857, SW1492, SW2021, SW2496 and SW2532) were used to genotype all samples (n=506). The set of markers and the amplification protocols are the same used by Scandura et al. (2008).
Data analysis
Raw data were checked for scoring errors due to stuttering or large allele dropout using the program MICRO-CHECKER 2.2.3 (van Oosterhout et al., 2004).
Previous results showed that a genetic discontinuity exists between the Italian peninsular and the other European populations (Scandura et al., 2008). Also, a genetic divergence was found between wild and domestic pigs. Accordingly, for assessment purposes the samples were grouped as follows: Sardinian wild boars (WSar), wild boars from the Italian peninsula (WIta), wild boars from the rest of Europe (WEur) and domestic pigs (DPig).
Allele frequencies and genetic diversity at the 10 loci were calculated for the four populations separately. Observed and expected (unbiased) heterozygosity, mean number of alleles per locus and FIS were computed in GENETIX v. 4.05 (Belkhir et al., 2001). Allelic richness was computed by using HP-RARE (Kalinowski, 2005) and by setting the rarefaction index to the minimum sample size.
Hardy–Weinberg equilibrium (HWE) and linkage equilibrium (LE) were tested in the Sardinian population using the software GENEPOP v. 3.4 (Raymond and Rousset, 1995). Tests for HWE employed the Markov chain method proposed by Guo and Thompson (1992), with the following chain parameters: 10 000 dememorizations, 100 batches and 5000 iterations. Deviations from LE were tested on genotypic data for each pair of loci. Significance levels were lowered, accounting for the number of multiple tests by the sequential Bonferroni procedure (Rice, 1989). Finally, the level of genetic differentiation between Sardinian wild boars and continental populations was estimated by calculating pairwise values of FST in GENETIX.
With the aim of investigating the genetic structure of the Sardinian population, we initially performed 10 independent Monte Carlo Markov chain runs in STRUCTURE v. 2.1 (Pritchard et al., 2000; Falush et al., 2003), simulating a number of subpopulations (K) ranging between 1 and 10. Simulations were performed using the admixture model and correlated allele frequencies, and consisted of 200 000 iterations after a burn-in period of 100 000 iterations. The optimal K-value was chosen according to the ΔK statistics developed by Evanno et al. (2005).
As translocations from continental populations were likely to have occurred in the past, in a further analysis we accounted for a gene flow from either Italian (that is, WIta) or other European (WEur) wild populations. In addition, a mix-up effect might arise from local crossbreeding with free-ranging domestic stocks (DPig).
Accordingly, we performed assignment tests to identify the origin of each sampled individual in the Sardinian population. Given that a subset of our data (292 individuals) had previously revealed a partition with four subpopulations, roughly corresponding to Sardinia, the Italian peninsula, the rest of Europe and domestic pigs (Scandura et al., 2008), we adopted two different Bayesian methods to assign Sardinian animals to one of these four clusters. The first method, implemented by the program BAPS v. 5.2 (Corander et al., 2008), uses stochastic optimization algorithms to identify populations with different allele frequencies and, under the mixture model, fully assigns an individual to one of the inferred clusters. The second Bayesian approach, employed by the software STRUCTURE v. 2.1, is based on Monte Carlo Markov chain randomizations of allele frequencies to determine the number of genetically distinct groups that fit HWE (Pritchard et al., 2000). In this case, under the admixture model, the algorithm calculates the individuals' membership proportion (Q) to each of the four inferred clusters.
We ran the mixture analysis in BAPS, setting K=4 (that is, forcing individuals to be grouped into a user-specified number of clusters equal to 4). In so doing, we expected that the clustering procedure would produce the same geographically coherent grouping as in Scandura et al. (2008). As this was the case in all 10 replicated runs, such an analysis allowed us to detect putative migrants in the inferred genetic mixture.
STRUCTURE was used to perform 10 independent Monte Carlo Markov chain simulations for K=4, disregarding any previous population information (settings were: admixture model, correlated allele frequencies, 100 000 iterations burn-in, 200 000 iterations data collection). The degree of admixture of sampled genomes was assessed by averaging the Q-values obtained during the 10 simulations. Individuals having >80% of membership to one cluster were assigned to the corresponding population. However, as individual Q-values varied among simulations, we took this variation into account by considering the standard deviation (s.d.) of the mean Q-value (Qm) as calculated across simulations. Accordingly, individuals were classified as ‘pure Sardinian’ if the corresponding Qm±s.d. for the Sardinian cluster was >0.8 (that is, if the range of variation of the Q-value as calculated across simulations fell entirely above the 0.8 threshold). Likewise, the individuals who averaged >0.8 membership (Qm±s.d.) to a cluster other than the Sardinian were classified as ‘recent immigrants’. Individuals were classified as ‘hybrids’ who had Qm±s.d. in the range of 0.2–0.8 for two (or more) clusters. All other individuals were classified as ‘unassigned’. We followed this approach to make our classification as conservative as possible by limiting a possible bias owing to differences among simulations.
To limit the effects of genetic introgression from alien sources on the pattern of population subdivision, a ‘purged’ dataset was created by including only wild boars classified as ‘pure Sardinian’ in the aforementioned STRUCTURE analysis.
Assuming the ‘purged’ Sardinian sample as representative of the native population, its consistency with HWE and LE, and the existence of a population structure were tested. After 300 000 Monte Carlo Markov chain iterations in STRUCTURE (starting with 100 000 iterations as burn-in), the convergence of Ln P(D) was reached for all simulations in the tested range K=1–10 (settings: admixture model, correlated allele frequencies, α inferred from an initial value of 1.0). The most likely number of subpopulations was assessed by the Evanno's method (Evanno et al., 2005).
Results
Microsatellite diversity of the overall Sardinian population
MICRO-CHECKER found no evidence of scoring errors in the dataset, whereas missing data amounted to 1.2% (0.5–3.3% per locus). The 10 microsatellite loci cumulatively showed 96 alleles in the Sardinian population (72% of the total number of alleles found across populations), with a range of 6–15 for each locus. The variability observed at each locus is shown in Table 1. In all loci, observed heterozygosity (Ho) was lower than expected (He), thus revealing an excess of homozygotes that was confirmed by FIS values and by significant deviations from HWE (P<0.01 for all loci; Table 1). Through comparing the overall microsatellite diversity of Sardinian wild boars with those found in continental populations and in domestic pigs (Table 2), we found very restricted ranges of He (0.611–0.657) and kAR (7.47–8.84), with surprisingly similar values for Sardinian and Italian peninsular wild boars. Moreover, private allelic richness (kPAR), which is independent from sample size, was higher in Sardinia than in mainland Italy (0.71). Tests of LE across the overall Sardinian population revealed that 10 out of 45 loci pairs (22%) were in disequilibrium at α=0.01 and 13 (29%) at α=0.05 after Bonferroni correction.
The estimated allelic divergence of the Sardinian population from continental populations, calculated as pairwise FST over loci (Table 3), was higher for the comparison WSar–WIta (0.091) and lower for WSar–WEur (0.063), while an intermediate level of differentiation was obtained from domestic pigs.
Analysis of population subdivision 1
The STRUCTURE analysis of the overall sample of Sardinian wild boars revealed a partition into five clusters (Figure 1a), even if the Evanno's method detected local maxima of ΔK values also at K=3 and 8. Three of the five inferred clusters had a geographically restricted distribution, one in Caprera island, one in north-west (NWS) and the other in south-west Sardinia (SWS), whereas the other two were spread in the northern and eastern part of the island (Figure 2a).
Results showing the most likely number of subpopulations (a) in the original and (b) in the ‘purged’ dataset of Sardinian wild boars. Dots represent the mean±s.d. of the log-likelihood (L(K)) associated with different values of K, calculated across 10 runs in STRUCTURE. Triangles connected by a solid line are values of ΔK calculated according to Evanno et al. (2005). The most likely partition is indicated by the arrow.
Genetic composition of the Sardinian wild boar population, obtained by the three-step Bayesian admixture analysis in STRUCTURE. Details of the computations are provided in the text. (a) Results of cluster analysis for 210 wild boars with K=5, in the absence of reference populations. Each individual is represented by a pie chart, in which colours represent the five inferred clusters. (b) Results of the assignment test for 210 individuals from Sardinia with K=4 and with a reference sample of Italian, European and domestic pigs. Individuals are represented by coloured dots, where colours express membership to a different cluster (light green—cluster I (Italian peninsula); blue—cluster II (continental Europe); red— cluster III (domestic pigs); yellow—cluster IV (Sardinia)). Individuals were classified as ‘pure Sardinian’ (full yellow), ‘recent immigrant’ (full red and full blue) or ‘hybrid’ (different colours) on the basis of their Q-values and of the consistency of these values across runs in STRUCTURE (see Figure 1). Wild boars that could not be classified on the basis of the adopted criteria are marked with a question mark and are coloured according to the cluster with the highest Q-value. (c) Results of the population admixture analysis performed on the ‘purged’ dataset (140 wild boars) using K=3 in STRUCTURE. Colours express membership to a different inferred cluster (sky-blue—cluster I; orange—cluster II; dark green—cluster III).
Detection of gene flow from other populations
The two Bayesian approaches, implemented by the programs BAPS and STRUCTURE, produced very similar results when used to group the 506 individuals in the dataset into four clusters (Figure 3). Both produced classification patterns that were consistent with the previous study (Scandura et al., 2008), and separated wild populations from peninsular Italy (cluster I), continental Europe (cluster II) and domestic pigs (cluster III) from Sardinian wild boars (cluster IV). The main difference concerned the assignment of wild boars sampled in France and Luxembourg. Although BAPS assigned them to the European cluster, admixture analysis in STRUCTURE recognized them as admixed between clusters II and III, and this result was consistent across simulation runs. In both analyses, all populations showed a certain number of possible immigrants (Figure 3).
Results of the two Bayesian analyses performed on 506 animals from wild boar and domestic pig populations across Europe. The upper graph refers to the cluster analysis performed with a stochastic optimization method in BAPS; by this approach each individual was fully (100%) assigned to the population where its alleles were most likely to occur. The lower graph is an admixture analysis performed in STRUCTURE, in which each individual is partitioned into the different clusters inferred by the program. In both cases, the number of subpopulations was set to K=4 and no population information was used to construct previous distributions. A full color version of this figure is available at the Heredity journal online.
On the whole, Bayesian clustering in BAPS assigned 176 (83.8%) out of 210 individuals from Sardinia to the Sardinian cluster (IV), 11 (5.2%) to peninsular Italy (I), 10 (4.8%) to continental Europe (II) and 13 (6.2%) to domestic pigs (III).
On the other hand, for most individuals the proportion of membership to the four inferred clusters in STRUCTURE was consistent across simulations (that is, Q-values had low s.d.; Figure 4). On the basis of Qm±s.d., five individuals (2.4%) were classified as ‘recent immigrants’. Three of them clustered with wild boars from continental Europe, whereas two of them were assigned to domestic pigs. These assignments were confirmed by the full match between BAPS and STRUCTURE results (Supplementary Figure S2). Two of the three continental wild boars were found in north-eastern Sardinia (Gallura), whereas the third was collected in the centre-west of the island (Monte Arci). The two domestic pigs were from the same area and were provided by the same hunter. The hypothesis of a trivial error in collecting/storing samples cannot be discarded. Under the same criteria, 140 individuals (66.7%) were classified as ‘pure Sardinian’, whereas 19 (9.0%) showed admixed ancestry (Figure 4 and Supplementary Figure S2). Seventeen cases of hybridization involved Sardinian wild boars: 10 implied crosses with WEur, seven with WIta and only one with domestic pigs, whereas in one case the admixture involved three components. The remaining two were WIta × WEur hybrids. Finally, 46 (21.9%) individuals could not be classified into one class, because of inconsistency across simulations (indicated by a question mark in Figure 2b and in Supplementary Figure S2). Ten specimens from Caprera island belonged to this group and were fully assigned to domestic pigs in BAPS, whereas their membership to the DPig cluster averaged >70% in STRUCTURE (Figure 3 and Supplementary Figure S2).
Assignment proportions to the four clusters (Qm±s.d., mean±standard deviations of the Q-values calculated over 10 independent simulations) estimated in STRUCTURE using the admixture model, correlated allele frequencies and simulating four different subpopulations (K=4). Samples from Caprera island were excluded (see the text). Colours represent the different inferred clusters (green—I; blue—II; red—III; black—IV). Individuals are classified as ‘pure Sardinian’, ‘recent immigrant’ or ‘hybrid’. The admixture zone includes individuals having a supposedly admixed ancestry. However, only those individuals with a conservative estimate of membership to two (or more) clusters were considered actual ‘hybrids’. A full color version of this figure is available at the Heredity journal online.
Analysis of population subdivision 2
To verify which effect the detected introgression has on the overall diversity and genetic structure of the population, we replicated the analysis in STRUCTURE omitting the 70 individuals who had been classified as either immigrants or hybrids, or non-confidently assigned to any category. Recalculated allele frequencies and estimates of genetic diversity revealed only a slight reduction of k (8.0) and He (0.602) with respect to the overall population (Table 4). As one would expect, recalculated pairwise FST-values revealed a higher divergence of the Sardinian population with respect to that estimated for the ‘non-purged’ Sardinian population (Table 3). Deviations from HWE and LE in the Sardinian population were still present after purging. However, the number of loci deviating from HWE decreased to 7 and the number of loci pairs in linkage disequilibrium decreased to 3 (6.7%, two of which were significant at α=0.01 and one at α=0.05), thus proving that genetic introgression contributed substantially to the disequilibria initially observed. On the other hand, the residual deviations were likely to be due to the observed genetic structure of the population.
This time the replicated Bayesian analysis in STRUCTURE identified three geographically consistent subpopulations. A peak of ΔK was indeed obtained for K=3, where the value of L(K) stabilizes (Figure 1b), but the occurrence of a local maximum at K=8 suggests a possible further substructure of the population. According to the main partition, a large portion of the island featured the presence of one major genetic cluster (east Sardinia, ES), whereas the two clusters, NWS and SWS, were fully confirmed (Figure 2c). Interestingly, it is exactly in these two areas that almost no signature of genetic admixture was found (Figure 2b). On the contrary, ES and in particular north-eastern Sardinia showed the highest proportion of genetic introgression from exotic populations (Figure 2b).
Allele frequencies in the three subpopulations did not deviate from HW expectations at any locus (Table 4). Analogously, no pair of loci showed a significant deviation from LE in any of them. Among the three subpopulations, SWS was the most and ES the least distant from exotic wild boar populations and from domestic pigs (Table 3). NWS showed an intermediate divergence from the other two areas (NWS–ES FST=0.128; NWS–SWS FST=0.119), which in turn showed a relevant differentiation (ES–SWS FST=0.182).
Discussion
Our results based on autosomal microsatellite data confirmed previous studies based on comparative morphometry and biochemical genetics (Groves, 1981; Apollonio et al., 1988; Randi et al., 1989; Randi, 1995), which were in accordance with the assessment of the peculiarity of Sardinian wild pigs with respect to all other wild boar populations. Thus, the historical classification of Sardinian pigs (together with Corsican pigs) as a different subspecies (Sus scrofa meridionalis) is now supported by a large amount of data. Microsatellites, like mitochondrial DNA (Randi, 1995; Scandura et al., 2008), revealed a high frequency of private alleles in the Sardinian population. Such a level of divergence could justify the status as an evolutionarily significant unit (Moritz, 1994; Fraser and Bernatchez, 2001), if isolation from other conspecifical populations were confirmed. However, the present investigation proves that the Sardinian population has lost its genetic isolation and has been genetically introgressed by imported continental wild boars that have admixed with Sardinian wild pigs. Approximately 11% of the individuals sampled throughout Sardinia were found to be either immigrants or hybrids with continental ancestors. These results were obtained in BAPS and STRUCTURE according to different iterative approaches, and were still consistent. The little difference between the outcomes of the two programs (for example, WSar075 and WSar158 in Supplementary Figure S2) can be attributed to the low power of the microsatellite set in discriminating between some European wild boars (France and Luxemburg) and the domestic pigs (Figure 3). Further sub-structuring with K=15 was reported by Scandura et al. (2008) for a similar dataset and can elucidate the aforementioned admixture showing the similarity between wild boars from France and domestic pigs (see Figure 6 therein).
The suspected genetic admixture of Sardinian wild boars with domestic pigs, which are free ranging in some areas of the island, was only marginally supported by our data. Had we excluded the isolated Caprera population, which resulted genetically closer to domestic pigs than to Sardinian wild boars and likely originated from a captive hybrid stock, and two individuals who were fully assigned to domestic pigs (possibly due to a sampling error), just one wild individual was ascertained to show the signature of hybridization with domestic animals. In our assessment of individual genetic purity, only hybridization events that occurred one to two generations before had a chance to be detected. Therefore, by virtue of the conservative approach that we used, the rate of hybridization is presumably underestimated.
Given this signature of genetic introgression from multiple sources, we wanted to test its effect on the genetic structure of the population. Our first analysis considered the complete Sardinian dataset (210 animals) and supported a clustering into five subpopulations. Two of them (NWS and SWS) were apparently unaffected by gene flow from exotic stocks and were confirmed in the second analysis, after discarding suspect immigrants and hybrids (including animals from Caprera island). In this case, however, the most likely partition was K=3, with a third subpopulation (ES) merging two clusters from the previous analysis. Genetic introgression was almost exclusively found in this subpopulation (Figure 2b).
Comparing the latter to the former analysis, if we exclude the cluster representing domestic pigs, the overall partition of the Sardinian wild population was reduced by one cluster. In other words, had we not considered the possible presence of introduced/introgressed individuals, this would have led to an overestimation of the population structure. Actually, as remarked by Bertorelle et al. (2009), depending on the source and number of displaced animals, an ‘artificial’ genetic structure can arise in the receiving population. An overestimation of the number of subpopulations may occur in two ways: by a misinterpretation of introgressed individuals as a different deme of native animals in the population or by affecting the performance of statistical programs. In the case of STRUCTURE, for instance, accounting for new (introgressed) alleles can affect the attempt to minimize deviations from HWE, thus leading to statistical solutions that differ from the original population structure.
NWS and SWS show a remarkable divergence from all non-Sardinian populations and from domestic pigs. These areas may have harboured relict wild boar nuclei during periods of demographic decline. Unfortunately, the lack of historical distribution and census data on the Sardinian population does not allow us to get insights into the time and duration of their isolation. In any case, the observed structure prompts an evaluation of the existence of possible cryptic barriers to gene flow in a landscape that apparently does not show physical obstacles to wild boar dispersal.
A remarkable genetic differentiation was revealed by Bayesian analysis also in other managed wild boar populations. In Portugal, Ferreira et al. (2009) showed a latitudinal partition into three subpopulations, and interpreted it as possibly originating during the last century from isolated refuge nuclei. Similarly, three to four clusters stand out in a transboundary population in central Europe, although isolation by distance seemed to concur to this pattern (Frantz et al., 2009). In that study, different clustering results were obtained depending on whether introduced individuals were included in the analysis or not. A striking differentiation was found between wild boars from the north and the south of Bulgaria, where the Thracian valley was reported to act as a barrier (Nikolov et al., 2009). Notably, in none of these studies the possibility of introgression from either exotic wild populations or local domestic stocks was taken into account.
Human-mediated gene flow is common in game species and can be a consequence of extensive restocking plans to recover harvested populations (DeYoung et al., 2003; Latch et al., 2006; Blanco-Aguiar et al., 2008) or of occasional translocations, which are often undocumented and/or illegal (Vernesi et al., 2003; Spencer and Hampton, 2005; Frantz et al., 2006, 2009; Crestanello et al., 2009). Unlike two other game species (Lepus capensis mediterraneus and Alectoris barbara), which were both historically introduced into the island and showed no or negligible signs of genetic introgression from continental stocks (Scandura et al., 2007, 2010), the Sardinian wild boar population shows the effects of a stronger extent of illegal introductions, mainly concerning the eastern part of the island. The strong recreational interest for this species, a negative demographic trend in Sardinia's past, the international trade opportunities and the ease with which wild boars can be raised in captivity may account for the present genetic composition of the population.
Nonetheless, in the light of our data, the Sardinian population should not be managed as a single panmictic unit. Rather, the three subpopulations (NWS, SWS and ES) should be treated as separate management units, particularly on account of the first two being important genetic stocks that show high levels of native diversity. Given its geographical location and degree of isolation, the SWS stock deserves special attention.
In conclusion, this study proves how a combination of molecular and statistical tools can help to discover cryptic patterns of natural variation and genetic structure while accounting for the effects of human perturbation. Here we stress the need to use appropriate reference samples in such analyses, including a range of possible source areas for translocated animals as well as an outgroup of the domestic population, wherever the possibility of hybridization with the domestic counterpart exists. Moreover, the possibility of an artificial overestimation of the number of clusters induces to be extremely cautious when making inferences on population structure in species that are strongly manipulated by human beings.
References
Albarella U, Manconi F, Rowley-Conwy P, Vigne J-D (2006). Pigs of Corsica and Sardinia: a biometrical re-evaluation of their status and history. In: Tecchiati U, Sala B (eds). Archaeozoological Studies in Honour of Alfredo Riedel. Province of Bolzano, Bolzano, Italy, pp 285–302.
Allendorf FW, England PR, Luikart G, Ritchie PA, Ryman N (2008). Genetic effects of harvest on wild animal populations. Trends Ecol Evol 23: 327–337.
Allendorf FW, Leary RF, Spruell P, Wenburg JK (2001). The problems with hybrids: setting conservation guidelines. Trends Ecol Evol 16: 613–622.
Apollonio M, Andersen R, Putman R (2010). Ungulate Management in Europe in the XXI Century. Cambridge University Press: Cambridge, UK.
Apollonio M, Randi E, Toso S (1988). The systematics of the wild boar (Sus scrofa L.) in Italy. Boll Zool 3: 213–221.
Belkhir K, Borsa P, Chikhi L, Raufauste N, Bonhomme F (2001). GENETIX 4.02, logiciel sous WindowsTM pour la génétique des populations. Laboratoire Génome, Populations, Interactions CNRS UMR 5000, Université de Montpellier II: Montpellier, France. Available at http://www.univ-montp2.fr/∼genetix/genetix/genetix.htm.
Bertorelle G, Papetti C, Hauffe HC, Boitani L (2009). Monitoring and detecting translocations using genetic data. In: Bertorelle G, Bruford MW, Hauffe HC, Rizzoli A, Vernesi C (eds.). Population Genetics for Animal Conservation. Cambridge University Press: Cambridge UK, pp 148–166.
Blanco-Aguiar JA, Gonzàlez-Jara P, Ferrero ME, Sànchez-Barbudo I, Virgós E, Villafuerte R et al. (2008). Assessment of game restocking contributions to anthropogenic hybridization: the case of the Iberian red-legged partridge. Anim Conserv 11: 535–545.
Boetticher H (1941). Zur Frage des sardinischen Wildschweines. Z Saugetierkd 14: 305–306.
Carta A, Usai MG, Sechi T, Casu S, Miari S, Porcu et al. (2007). Genetical variability of the Sardinian pig population at microsatellite loci. Sixth International Symposium on the Mediterranean Pig, 2007: Capo d'Orlando, Italy.
Choisy M, Franck P, Cornuet M (2004). Estimating admixture proportions with microsatellites: comparison of methods based on simulated data. Mol Ecol 13: 955–968.
Corander J, Marttinen P, Sirén J, Tang J (2008). BMC Bioinformatics in BAPS software for learning genetic structures of populations. BMC Bioinform 9: 539.
Corander J, Waldmann P, Sillanpaa MJ (2003). Bayesian analysis of genetic differentiation between populations. Genetics 163: 367–374.
Crestanello B, Pecchioli E, Vernesi C, Mona S, Martinkova N, Janiga M et al. (2009). The genetic impact of translocations and habitat fragmentation in Chamois (Rupicapra) spp. J Hered 100: 691–708.
DeYoung R, Demarais S, Honeycutt RL, Rooney AP, Gonzales RA, Gee KL (2003). Genetic consequences of white-tailed deer (Odocoileus virginianus) restoration in Mississipi. Mol Ecol 12: 3237–3252.
Evanno G, Regnaut S, Goudet J (2005). Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol Ecol 14: 2611–2620.
Falush D, Stephens M, Pritchard JK (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Ferreira E, Souto L, Soares AMVM, Fonseca C (2009). Genetic structure of the wild boar population in Portugal: evidence of a recent bottleneck. Mamm Biol 74: 274–285.
Frantz AC, Cellina S, Krier A, Schley L, Burke T (2009). Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance? J Appl Ecol 46: 493–505.
Frantz AC, Tigel Pourtois J, Heuertz M, Schley L, Flamand MC, Krier A et al. (2006). Genetic structure and assignment tests demonstrate illegal translocation of red deer (Cervus elaphus) into a continuous population. Mol Ecol 15: 3191–3203.
Fraser DJ, Bernatchez L (2001). Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol Ecol 10: 2741–2752.
Groves C (1981). Ancestors for the Pigs: Taxonomy and Phylogeny of the Genus Sus. Technical Bulletin, Research School of Pacific Studies, Australian National University: Canberra, Australia.
Guillot G, Estoup A, Mortier F, Cosson JF (2005). A spatial statistical model for landscape genetics. Genetics 170: 1261–1280.
Guo SW, Thompson EA (1992). Performing the exact test of Hardy–Weinberg proportions for multiple alleles. Biometrics 48: 361–372.
Kalinowski ST (2005). HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5: 187–189.
Latch EK, Harveson LA, King JS, Hobson MD, Rhodes Jr OE (2006). Assessing hybridization in wildlife populations using molecular markers: a case study in wild Turkeys. J Wildl Manage 70: 485–492.
Lecis R, Pierpaoli M, Biro ZS, Szemethy L, Ragni B, Vercillo F et al. (2006). Bayesian analyses of admixture in wild and domestic cats (Felis silvestris) using linked microsatellite loci. Mol Ecol 15: 119–131.
Major F (1883). Studien zur Geschichte der Wildschweine (Gen. Sus). Suppl Zoologischen Anzeiger 140: 295–300.
McDevitt AD, Edwards CJ, O'Toole P, Sullivan P, O'Reilly C, Carden RF (2009). Genetic structure of, and hybridisation between, red (Cervus elaphus) and sika (Cervus nippon) deer in Ireland. Mamm Biol 74: 263–273.
Moritz C (1994). Defining evolutionary significant units for conservation. Trends Ecol Evol 9: 373–375.
Nikolov IS, Gum B, Markov G, Kuehn R (2009). Population genetic structure of wild boar Sus scrofa in Bulgaria as revealed by microsatellite analysis. Acta Theriol 54: 193–205.
Oliver WLR, Brisbin IL, Takahashi S (1993). The Eurasian wild pig, Sus scrofa. In: Oliver WLR (ed). Pigs, Peccaries and Hippos—Status Survey and Action Plan. IUCN/SSC, Gland: Switzerland, pp 112–121.
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Randi E (1995). Conservation genetics of the genus Sus. Ibex 3: 6–12.
Randi E, Apollonio M, Toso S (1989). The systematics of some Italian populations of wild boar (Sus scrofa L.): a craniometric and electrophoretic analysis. Z Säugetierkd 54: 40–56.
Raymond M, Rousset F (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249.
Rice WR (1989). Analyzing tables of statistical tests. Evolution 43: 223–225.
Scandura M, Iacolina L, Apollonio M, Dessì-Fulgheri F, Baratti M (2010). Current status of the Sardinian partridge (Alectoris barbara) assessed by molecular markers. Eur J Wildl Res 56: 33–42.
Scandura M, Iacolina L, Ben Slimen H, Suchentrunk F, Apollonio M (2007). Mitochondrial CR-1 variation in Sardinian hares and its relationships with other Old World hares (Genus Lepus). Biochem Genet 45: 305–323.
Scandura M, Iacolina L, Crestanello B, Pecchioli E, Di Benedetto F, Russo V et al. (2008). Ancient vs. recent processes as factors shaping the genetic variation of the European wild boar: are the effects of the last glaciation still detectable? Mol Ecol 17: 1745–1762.
Simberloff D (1996). Hybridization between native and introduced wildlife species: importance for conservation. Wildl Biol 2: 143–150.
Spencer PBS, Hampton JO (2005). Illegal translocation and genetic structure of feral pigs in Western Australia. J Wildl Manage 69: 377–384.
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4: 535–538.
Vernesi C, Crestanello B, Pecchioli E, Tartari D, Caramelli D, Hauffe H et al. (2003). The genetic impact of demographic decline and reintroduction in the wild boar (Sus scrofa): a microsatellite analysis. Mol Ecol 12: 585–595.
Wilkens B (2003). La fauna sarda durante l'Olocene: le conoscenze attuali. Sardinia, Corsica et Baleares Antiquae I: 181–197.
Acknowledgements
We thank A Cois, P Onida, A Pintore, H Okarma, L Sugar, J Carranza, F Suchentrunk, E Baubet, G Kaminski, L Schley, R Mazzoni Della Stella, S Antonacci, P Varuzza, L Tonini, the Istituto Superiore per la Protezione e Ricerca Ambientale (ISPRA) and several hunters and pig breeders for assisting us in sample collection. We are grateful to E Pecchioli and B Crestanello for sharing molecular data. We also thank A Frantz, G Bertorelle and S Ciuti, who provided valuable comments to the manuscript, and BA Brandon for linguistic revision. This research was financially supported by the Sardinian Regional Government (LR 7/2007 Promozione della Ricerca Scientifica e dell'Innovazione Tecnologica in Sardegna) and by the Provincial Administration of Cagliari.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Scandura, M., Iacolina, L., Cossu, A. et al. Effects of human perturbation on the genetic make-up of an island population: the case of the Sardinian wild boar. Heredity 106, 1012–1020 (2011). https://doi.org/10.1038/hdy.2010.155
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2010.155
Keywords
This article is cited by
-
Assessing population structure and migration patterns of wild boar (Sus scrofa) in Japan
Scientific Reports (2023)
-
Spatial genetic structure of European wild boar, with inferences on late-Pleistocene and Holocene demographic history
Heredity (2023)
-
Genetic characterization and implications for conservation of the last autochthonous Mouflon population in Europe
Scientific Reports (2021)
-
Matching STR and SNP genotyping to discriminate between wild boar, domestic pigs and their recent hybrids for forensic purposes
Scientific Reports (2020)
-
Hotspots of recent hybridization between pigs and wild boars in Europe
Scientific Reports (2018)