Abstract
The measurement of telomere length (TL) is a genetic tool that is beginning to be employed widely in ecological and evolutionary studies as marker of age and fitness. The adoption of this approach has been accelerated by the development of telomere quantitative PCR, which enables the screening of large numbers of samples with little effort. However, the measurement and interpretation of TL change need to be done with a necessary level of rigour that has thus far often been missing where this approach has been employed in an ecological and evolutionary context. In this article, we critically review the literature available on the relationship between TL, age and fitness. We seek to familiarize geneticists, ecologists and evolutionary biologists with the shortcomings of the methods and the most common mistakes made while analysing TL. Prevention of these mistakes will ensure accuracy, reproducibility and comparability of TL studies in different species and allow the identification of ecological and evolutionary principles behind TL dynamics.
Similar content being viewed by others
Introduction
Telomeres, the ends of linear chromosomes, comprise a repetitive DNA sequence (Meyne et al., 1989) and associated proteins (shelterin, de Lange, 2005). Telomeres are important for proper positioning of chromosomes during replication (Kirk et al., 1997), they can reversibly repress the expression of subtelomeric genes (Mefford and Trask, 2002) and they protect the chromosome end from being recognized as DNA double-strand breaks by the DNA damage response machinery (de Lange, 2002).
Owing to the end replication problem and other factors (Bolzan and Bianchi, 2006), telomeres shorten with every cell division if not restored by telomerase (Aubert and Lansdorp, 2008) or alternative telomere lengthening pathways (Bolzan and Bianchi, 2006). If telomere length (TL) falls below a certain threshold, the cell enters senescence or apoptosis (Bolzan and Bianchi, 2006). It is therefore believed that telomeres are part of a mechanism to detect the number of cell replications and DNA damage with the function of preventing uncontrolled cell proliferation, the main hallmark of cancer (Wright and Shay, 2005; Aubert and Lansdorp, 2008). On the other hand, while protecting the body from cancer, TL-induced senescence also reduces the body's ability to regenerate damaged or worn out cells, which is believed to be one of the reasons why most animals age and show an age-related increase of mortality (Wright and Shay, 2005).
There is a large amount of literature on the measurement of TL in humans, with numerous correlations to age and lifestyle factors that accelerate ageing and morbidity (Aviv, 2006; Baird, 2006, 2008). However, it is only relatively recently that TL has been adopted as a molecular marker for estimating age and fitness in ecological and evolutionary context (Haussmann and Vleck, 2002; Nakagawa et al., 2004; Monaghan and Haussmann, 2006; Bize et al., 2009). To date, most of these studies have focused on bird species (Hall et al., 2004; Haussmann et al., 2005; Juola et al., 2006; Pauliny et al., 2006; Bize et al., 2009; Salomons et al., 2009), and a few on reptiles (Scott et al., 2006; Hatase et al., 2008) and fish (Hatakeyama et al., 2008; Horn et al., 2008; Hartmann et al., 2009).
In his review of the use of TL in human epidemiology, Aviv (2008) showed that there was ‘widespread confusion,… rooted in conflicting findings with opposing conclusions’ and that ‘the rush to publish has often won out over adherence to epidemiological principles’. Although the use of TL change in ecology and evolutionary biology is in its infancy when compared with its use in humans, we need to heed similar warnings on the use of this methodology to avoid generating similar problems and errors in analysis and interpretation. Such warnings are timely with the increasing use of TL in ecological and evolutionary studies. The development of telomere real-time quantitative (Q)-PCR (Cawthon, 2002, 2009) also threatens the integrity of telomere dynamics as a tool in ecology. Although the ease with which results may be obtained using this method is alluring, the multiple control and optimization reactions required to conduct a reliable Q-PCR (Bustin and Nolan, 2004; Bustin and Mueller, 2005; Aviv, 2008) are less so, and consequently often omitted from the experimental procedures (Aviv, 2009).
Here, we critically review the results of published studies that have investigated TL in relation to age and fitness and the misconceptions, we believe, have been generated and promoted as a result of inappropriate methodology or data interpretation. We then focus on the two main methods used to measure TL, telomere restriction fragment (TRF) analysis and telomere Q-PCR, highlighting mistakes and omissions that have been made in the past. As this is a new field, we do not wish to imply any fault by researchers or research groups who have pioneered the use of TL in an ecological and evolutionary context. Rather, our intention is to use the collective experience gained in these past few years from our laboratory and others, alongside the growing data available from research on humans, to point out the shortcomings of the currently available approaches for measuring TL in an ecological and evolutionary context so that these mistakes may be avoided in future research.
TL and age
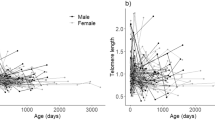
The concept of telomere ageing was introduced into ecology and evolutionary biology by Haussmann and Vleck (2002) and further promoted by Haussmann et al. (2003b). The authors found a significant correlation between TL and age in a variety of bird species and suggested the use of TL as a tool to estimate the age of individuals. As the knowledge of a population's age structure is one of the most crucial factors influencing interpretation of life history parameters (Haussmann and Vleck, 2002; Monaghan and Haussmann, 2006), the idea of estimating age by measuring TL has great appeal and was popularized further (Nakagawa et al., 2004) and tested by others (Juola et al., 2006; Hatase et al., 2008). Although theoretically there is potential to obtain age from TL, most authors have limited themselves to simply examining the correlation between TL and age. To date, there are only two studies that actually tried to use TL to investigate the age of unknown individuals: Juola et al. (2006) and Hewakapuge et al. (2008).
Hewakapuge et al. (2008) tested whether TL measured by telomere Q-PCR could be used to determine the age of an unknown human in forensic investigations. The authors found a correlation between TL and age, but the coefficient of determination (R2) was so low that age based on TL could not even be approximated. The authors concluded that telomere ageing does not work in humans because of high variability of TL between age-matched individuals, presumably due to genetic and environmental differences (Hewakapuge et al., 2008).
The second study investigated the potential of telomere ageing in an ecological framework (Juola et al., 2006). The authors used 36 frigatebirds (Fregata minor) of known age to construct a calibration curve for ageing unknown birds from the same population. Using a 95% confidence interval, the authors were unable to achieve a reliable assignment of age for the unknown individuals. For example, the smallest 95% confidence interval in this study had a range of 36.4 years and some birds were assigned to an age class that ranged between 72 and 205 years, despite the maximum reported lifespan of this species being 44 years (Juola et al., 2006). The authors concluded that frigatebirds do not fulfil the requirements necessary for telomere ageing, namely, large variation of TL with time and little variation in initial TL (Nakagawa et al., 2004). In this context, it is important to note that this study found the highest R2 (0.82) ever reported between age and TL in birds (Table 1). If this species is not suitable for telomere ageing, it may be difficult to find any species that will be.
There are several reasons why telomere ageing has not been successful to date and indeed might never be. One reason is that the methodologies used to measure TL often have some degree of arbitrariness in terms of interpretation (for example, start and end of TRF smear, see below), which impedes reproducibility. Some attempts have been made to remove the arbitrariness of measurement by suggesting general guidelines for TL analysis (for example, telomere optimal estimate; Haussmann and Mauck, 2008) or the use of universal standard samples (Aviv, 2008). These are reviewed below.
We believe, however, that the main reasons why ageing based on TL change may ultimately be unsuccessful are biological in nature. First, many studies have found that the rate of TL change varies over time in both birds (Hall et al., 2004; Juola et al., 2006; Pauliny et al., 2006; Salomons et al., 2009) and other animals (Frenck et al., 1998; Brummendorf et al., 2002; Baerlocher et al., 2007). Often a high rate of TL change is observed early in life that then gradually lessens throughout life, before a possible acceleration shortly before death (Salomons et al., 2009). A calibration curve for ageing based on TL measurements for only a few individuals is unlikely to account for this change in rate of telomere loss throughout life, biasing estimates accordingly. Second, all studies to date relating TL and age show high variability in TL between age-matched individuals. Although a general trend of TL change can still be measured in many species, a reliable age estimation for individual animals is impossible if TL within an age group spans ⩾80% of the total TL range of the data set (for example, Haussmann et al., 2003b; Hall et al., 2004; Pauliny et al., 2006; Bize et al., 2009). It is important not to confuse correlation with predictability. A coefficient of determination of 0.5 or even more is a very convincing result when analysing relationships between two factors on an ecological or evolutionary scale, but is not enough to accurately estimate the age of individuals. Even a coefficient of determination of 0.82 was not enough to estimate age in the frigatebird (Juola et al., 2006).
Finally, there are some species that seem to show no change of TL with age, at least in adult individuals (Hall et al., 2004; Pauliny et al., 2006). Some species seem to have evolved mechanisms to avoid erosion of telomeres in adulthood and do not show any decline of TL with age without having an increased risk of cancer (Hall et al., 2004). Such species will be resistant to any attempt to estimate age from TL.
TL and fitness
As short telomeres induce senescence in cells and hence reduce the regenerative capacity of the corresponding tissues, it has been suggested that TL might be correlated with various fitness parameters (Nakagawa et al., 2004; Monaghan and Haussmann, 2006). In fact, TL has been linked to survival and reproductive success in some bird species (Haussmann et al., 2005; Pauliny et al., 2006; Bize et al., 2009).
However, age is also correlated with survival and reproductive success, and thus TL has to be corrected for age in mixed cohorts to give the residual TL (rTL) (Pauliny et al., 2006), which indicates whether individuals have shorter or longer TLs than expected for their age. Unfortunately, the differences in rTL among individuals that are correlated with fitness are inevitably very small (Pauliny et al., 2006) and hence methodologically difficult to measure. Alternatively, TL and fitness can be compared in age-matched groups between quartiles of TL rather than directly correlated with TL by regression (Haussmann et al., 2005; Bize et al., 2009).
Haussmann et al. (2005) reported a five times higher probability for first-year tree swallows (Tachycineta bicolor) of the top quartile for TL to return to the breeding ground the next year than for the birds of the bottom quartile for TL. However, these results might be biased by the inclusion of values obtained by two different methodological protocols used for old and new samples (Haussmann et al., 2005). Unfortunately, it is not clear what exactly the difference between the methods is. The use of pulse field electrophoresis instead of constant field electrophoresis alone cannot explain the different magnitudes of estimated TL. Most likely the authors used the later described telomere optimal estimate method (Haussmann and Mauck, 2008, see below) for the new samples, although this method has been shown to yield different results for TL in both magnitude and distribution among individuals (Haussmann and Mauck, 2008). In this case the two data sets should not be combined. Therefore, the relationship reported between TL and survival in tree swallows needs further scrutiny and, ideally, independent confirmation.
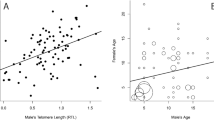
Pauliny et al. (2006) investigated the relationship between rTL and fitness in sand martins (Riparia riparia) and dunlins (Calidris alpine). Although in sand martins rTL was positively correlated with minimum lifespan (estimated by the last date the birds were seen), it was not in dunlins. In dunlins, rTL was negatively correlated with tarsus length. Furthermore, rTL was positively correlated with the categorical measure of whether male dunlins produced offspring during their lifespan or not. As smaller male dunlins are preferred by female dunlins, rTL was indirectly (via body size) and directly (via reproductive success) associated with evolutionary fitness in male dunlins.
Taken together with the lifespan correlation in sand martin, the authors suggested that rTL could be used as a measure for fitness in both species. Although such results support a link between TL and fitness, one needs to keep in mind that selected fitness measures were correlated only in different species and sex. Lifespan was only correlated with rTL in sand martins, but not in dunlins. Tarsus length and recruitment of offspring was only correlated with rTL in male dunlins and not in females. Furthermore, the number of offspring of male dunlins did only show a non-significant association with rTL. Although these findings support a connection between TL and fitness, they also show that these correlations are highly specific and cannot be used as a general marker in ecological studies on species not studied before.
Bize et al. (2009) found that in Alpine swift (Apus melba), TL and telomere rate of change (TROC) were a predictor of survival, whereas chronological age was not. A combination of long telomeres and slow erosion rate gave the highest probability for survival. The fact that TROC was a predictor of survival independent of actual TL shows that TL is just one factor linking telomere dynamics to lifespan and care should be taken when using only TL as a marker for survival and fitness.
Recently, Salomons et al. (2009) reported that individuals with shorter TL are more likely to disappear from the population and that this disappearance is preceded by rapid loss of telomere sequences. However, as TL differs between two figures in the paper that reportedly show the same data set (cf. Figures 2a and b), there is some doubt around the analytical procedure leading to this finding. This uncertainty highlights the importance of describing methods of data analysis as accurately as the method of TL measurement.
In summary, there is theoretical and experimental evidence for a connection between TL and fitness parameters. However, TL differences are small and species and/or sex specific. Only survival seems to be linked to TL in a number of species and hence might be useful as a general marker in ecological and evolutionary studies. However, recent studies suggest that this effect might be detectable only shortly before death in humans and birds (Baird, 2006; Salomons et al., 2009). Within species TL might be an appropriate measure for selected parameters of fitness, but it remains to be seen whether it is more feasible to measure TL as a surrogate of these parameters or to directly measure these parameters themselves.
TL and environment
The influence of the environment on TL and TL change has been widely investigated in humans (Aviv, 2006; Baird, 2006, 2008), but little is known about other species under natural conditions. One study investigated the effect of intrinsic and extrinsic factors early in life on the TROC in European shag (Phalacrocorax aristotelis) (Hall et al., 2004). They found that the TROC was correlated with TL as a chick, body mass corrected for size and date of egg laying, and that together all three factors could explain 61% of the variation in TROC observed in shag. Consequently, TL and individual fitness might be connected via a feedback loop that accelerates into a downward spiral in the late stages of life. Indeed, this might explain the apparent observation of a rapid decline of TL and fitness towards the presumed end of a bird's life (Salomons et al., 2009).
In the European sea bass (Dicentrarchus labrax), different age classes tend to have distinct ranges of TL, although TL did not correlate with age (Horn et al., 2008). The authors suggested that environmental factors might have a role in telomere dynamics, as all individuals of one age class were reared under the same conditions. However, a common genetic basis of hatchery stocks might also explain the pattern found. Environmental factors, interacting with an underlying genetic predisposition to telomere maintenance or loss, may influence telomere dynamics in this fish and probably in other species. Increasingly this interaction has been observed in humans with TL reduction being linked to factors, such as oxidative damage (Von Zglinicki, 2002), stress (Epel et al., 2004), smoking (Valdes et al., 2005), diet and growth (Demerath et al., 2004), exercise (Cherkas et al., 2006) and blood pressure (Demissie et al., 2006), while also being heritable (Baird, 2006, 2008; Njajou et al., 2007).
Methods for TL measurement
Given the high conservation of telomere sequence, telomere-associated proteins and pathways leading to senescence (Meyne et al., 1989; de Lange, 2002; Traut et al., 2007), one would assume that TL and its regulation is similar among related species and diverges with phylogenetic distance. However, a broad comparison of telomere dynamics across taxa is hindered by methodological differences between studies. In this section, we discuss how variances within and differences between the methods used to measure TL (namely, TRF and telomere Q-PCR) can hinder meta-analysis of TL data and what can be done to minimize these differences.
In this review, we do not discuss fluorescence in situ hybridization (FISH) or flow-FISH, as these methods are rarely employed by ecologists. The main disadvantage of FISH is that it usually requires dividing cells displaying metaphase spreads (Londono-Vallejo et al., 2001), which are almost impossible to obtain in ecological field studies. However, FISH could help identify ultra-short telomeres and interstitial telomeric repeats if dividing cells can be obtained from selected individuals. Flow-FISH, a combination of FISH and flow cytometry is capable of rapidly and accurately measuring the TL of different blood cells (Rufer et al., 1999), and shows some potential to be used in ecological studies. Unfortunately, the utility of Flow-FISH is hampered by the need for highly specialized equipment and training and thus has so far only been used in human and baboon (Baerlocher et al., 2007).
TRF analysis
TRF analysis is the traditional method to measure TL and is still considered the gold standard (Aviv, 2009). Genomic DNA is cut by restriction enzymes to obtain telomeric restriction fragments that are resolved by gel electrophoresis and detected by hybridization (Harley et al., 1990). As all telomeres have different lengths, a telomere smear of telomeric fragments rather than a distinct band is obtained and it is this smear that is used to calculate the mean TL. Although TRF is often referred to as one method to measure TL, it actually is a general term for a variety of different experimental and analytical procedures. These can differ in restriction enzymes, hybridization targets (the whole telomere vs overhang), hybridization probes (radioactive vs chemiluminescence), hybridization conditions (in-gel vs Southern blot), gel calibration, background subtraction, the formula used to calculate TL and the analysis window selected. Each one of these factors can influence TRF analysis and therefore should be accounted for when studies of TL are compared, particularly when the comparison is among different species.
The first step when analysing TRF images is the estimation of the fragment size range against molecular weight markers. In general, migration of DNA in agarose gels follows a log-linear distribution within a defined range (Sambrook and Russell, 2001). However, if the analysis window is larger than this range, exponential regression might not be appropriate to calibrate the gel and can lead to distortions in the size distributions (Horn, 2009). The differences introduced between studies by gel calibration might seem trivial, as all regressions have high r-values, but if only subsets of the distribution are analysed, as suggested by Haussmann and Mauck (2008), this factor can become a significant obstacle when comparing TL and TL change between studies. In addition, a reliable and reproducible TL estimate is questionable in studies that use an analysis window that entirely lies outside the actual range of the molecular weight markers (for example, Haussmann and Mauck, 2008).
Another factor influencing TL estimates is the analysis program and/or formula used to calculate the mean TL. The most frequently used formulae are

and

where ODi is the optical density at position i; and MWi is the molecular weight at position i. Equation (1) calculates the geometric mean of the distribution and is used for non-denaturing blots in which the telomere signal is similar for each telomere as the probe is only bound to the single-stranded overhang of the telomeres. The second formula is used for denaturing blots and corrects for increased binding of probes to longer telomeres (Grant et al., 2001). The program telometric (Grant et al., 2001) has also often been used to calculate TL (Juola et al., 2006; Pauliny et al., 2006; Horn et al., 2008), but has been criticized for yielding biased results (Haussmann and Mauck, 2008; Salomons et al., 2009) and being known to use an incorrect algorithm (Salomons et al., 2009). However, our own experience with telometric and our review of the published literature in which telometric has been used suggests that there is no support for the claim that it uses an incorrect algorithm (Horn et al., 2008; Horn, 2009). Finally, the position of the peak of the telomere distribution has also been used as a measure for mean TL (Londono-Vallejo et al., 2001; Cottliar et al., 2006).
Unfortunately, no study to date has investigated whether the use of these different formulae leads to similar TL estimated for both non-denaturing and denaturing blots. It is clear, however, that the choice of program or formula can significantly alter the output data. How the variance among these formulae might affect comparisons must be considered. What is more there is the obvious temptation for investigators to simply utilize the formula or approach that gives rise to the pattern that most closely matches the pattern expected or desired, hence firmer guidelines for best practice in such analyses are needed.
Probably the parameter in TRF that varies most from study to study is the analysis window. Traditionally the whole telomere smear from a TRF blot has been used to estimate mean TL (Harley et al., 1990; Hastie et al., 1990). Indeed, when Haussmann and Vleck (2002) introduced telomere dynamics to the field of ecology using zebra finch, they used the range of 3–17 kb introduced by Harley et al. (1990). However, in contrast to the smear of human telomeres measured by Harley et al. (1990), zebra finch telomeres range well above 17 kb, consequently only a part of the telomere signal was analysed. In 2003, contradicting methods were reported in two separate papers based on the same data set; an analysis window of 2–13 kb was reported for common terns (Sterna hirundo) in Hausmann et al. (2003a), whereas this window increased to 3–30 kb in Haussmann et al. (2003b). Other authors subsequently used the whole telomere distribution (Hall et al., 2004), an analysis window that included most of the distribution (Pauliny et al., 2006) or a window from the bottom of the telomere smear to just below the limit of mobility (Haussmann et al., 2003a; Juola et al., 2006). However, in the later approach the limit of mobility is rarely defined, which makes comparisons among studies, even in the same species, challenging.
Haussmann and Mauck (2008) attempted to address the uncertainty in defining the analysis window. They suggested focusing on the shortest telomeres and presented a method to identify the analysis window (telomere optimal estimate), which was found to show the best correlation between TL and age out of hundreds of analysis windows tested in their study. Of course this approach assumes that a correlation between TL and age exists for the species being studied and then tries to find/maximize this correlation in a large number of data sets produced using different analysis windows. Although this strategy may be a legitimate approach to optimize the correlation between TL and age for the purpose of ageing animals, any additional biological meaning applied to this estimate is questionable.
Finally, Salomons et al. (2009) defined the upper limit of the analysis window based on the peak intensity and the intensity at the side of the distribution. This approach was necessary because of high background noise at the upper end of the TRF distribution. This method seems to be specific for each lane and consequently might be susceptible to bias due to differences in DNA quantities between samples. In our experience, high background intensities suggest the TRF method has not been fully optimized and should be avoided experimentally during hybridization and detection rather than eliminated mathematically leading to a truncation of the TRF distribution.
Telomere Q-PCR
Telomeres were thought to be impossible to amplify by PCR because of their repetitive nature until Cawthon (2002) presented a cleverly designed primer set. He introduced several mismatches to prevent annealing of two primers to each other, but that still allow annealing to the telomeric sequence to facilitate PCR amplification. The amount of amplified DNA measured by telomere real-time PCR (or Q-PCR) is proportional to the amount of initial template (that is, telomeric sequence). The initial quantity of telomeric sequences depends on the DNA concentration and on the TL. To control for DNA concentration, a single-copy gene (SCG), 36B4 in the initial study (Cawthon, 2002) is amplified in a separate reaction and the ratio of telomere and SCG concentration (T/S) is expressed relative to a reference sample. This ratio is proportional to TL as shown by correlation with TRF analysis (Cawthon, 2002).
The ease with which results can be produced using real-time PCR, its high-throughput capacity and its low costs will probably lead to telomere Q-PCR becoming the method of choice over TRF. However, several factors can confound reliability of Q-PCR and these need to be carefully controlled. Potentially, the most important factor is amplification efficiency (Nordgard et al., 2006). Despite this, a number of publications in human and other species have either omitted amplification efficiency (Epel et al., 2006; Hartmann et al., 2009) or even present amplification efficiency data that prove their results unreliable (Zhang et al., 2007; discussed in Horn, 2008). Another problem is the choice of the SCG required for telomere Q-PCR. For example, a well-known multi-locus gene (as used in Hatase et al., 2008) is not an acceptable control gene for telomere Q-PCR.
Many examples of measurement errors arising from methodological shortcomings of telomere Q-PCR are from the field of epidemiology, but we believe that it is important to bring these errors to the attention of researchers who want to use this method in an ecological or evolutionary setting so that they may rapidly learn from mistakes made in other disciplines. If these mistakes are copied over to ecology from publications in other fields, we fear that this will create a level of confusion that exceeds that already noted in epidemiology by Aviv (2008, 2009).
The most important factor affecting a real-time PCR assay is inhibition of the reaction and the variation in amplification efficiency that results (Pfaffl, 2001; Nordgard et al., 2006). Efficiency is the measure of how much of the target sequence is amplified in each cycle. An efficiency of 100% is defined as a doubling of target sequence in each PCR cycle (sometimes it is also defined as 2, where efficiency in percentage (x%) can be converted to the decimal number (y) using the equation y=x%/100+1; Rebrikov and Trofimov (2006)). As real-time quantification calculates from a threshold cycle (Ct) back to the initial amount of template DNA in the reaction (cycle 0), it is important that the amplification efficiency is the same for all samples. What might be considered small deviations in efficiency matter greatly—a deviation of 5% efficiency between the standard and the sample can lead to a measurement error of 88% if the sample is detected after 25 cycles (for details see Supplementary Table S1).
Telomere real-time PCR is especially vulnerable to efficiency errors, because two independent amplifications (telomere and SCG) are used to estimate the relative TL, each of which will probably include efficiency errors. Different thresholds for acceptable efficiencies have been suggested (for example, 90–110% (Stratagene, 2004) and 93–105% (Nolan et al., 2006)). These limits are only indicative, as the actual values are of minor importance if all samples show the same efficiency. Quantification with all reactions having an efficiency of 80% is as reliable as all samples having 100% because only the differences between efficiencies lead to efficiency error. Unfortunately, most studies do not check the efficiency of their samples, and it is simply assumed that there is a higher chance of consistency between the standard and sample efficiency if the standard is optimized to a value around 100% (the theoretical perfect amplification). This, however, is a very dangerous assumption (Horn, 2009). A variety of chemicals used in standard blood storage and DNA extraction methods inhibit PCR (for example, heparin, SDS, phenol, ethanol, EDTA; Wilson, 1997) and might be present in different concentrations in DNA extractions from similar biological materials.
Out of 21 randomly selected studies using telomere Q-PCR (see Horn, 2009), only two mention the amplification efficiency of their standard curves (Gil and Coetzer (2004) and Criscuolo et al. (2009)). Two others show a figure including the slope of the standard curves (Nordfjall et al., 2005; Zhang et al., 2007) that can be used to deduce efficiency. Cawthon (2002) indicated an efficiency of 80–100% on request (H Thorsten, personal communication), but our experience is that most studies do not mention amplification efficiency and thus simply cannot be evaluated.
Although the efficiencies obtained by Gil and Coetzer (2004), Nordfjall et al. (2005), Criscuolo et al. (2009) and perhaps that of Cawthon (2002) were in an acceptable range, the amplification efficiency of the telomere reaction reported in other studies is far beyond acceptability (for example, Zhang et al., 2007). It is also worrying that a study investigating the effects of tissue-fixing agents (for example, formaldehyde or RNAlater) on telomere Q-PCR (Koppelstaetter et al., 2005) completely ignores the issue of amplification efficiency, although these fixatives are obligatory PCR inhibitors as they are designed to stop any cellular enzyme activity, including polymerases.
Another problem with telomere Q-PCR amplification efficiency is introduced by the use of external standards as a reference sample (Martin-Ruiz et al., 2004; Grabowski et al., 2005; Fehrer et al., 2006; O'Callaghan et al., 2008). The use of a reference sample, which should generally be one of the test samples, or a pool of many samples, is assumed to at least partly control for the mixture of PCR inhibitors present in any clinical DNA sample. Thus, optimization of the reaction conditions using this sample increases the chance of minimizing amplification efficiency differences between samples and reference. In contrast, a reaction optimized to a highly purified commercial sample (Martin-Ruiz et al., 2004) or DNA extracted for a cell line (Grabowski et al., 2005; Fehrer et al., 2006) completely ignores any PCR inhibitor present in the clinical samples. Furthermore, additional inhibitors can be introduced by different cell lines, as apparent from the different slopes of the standard curves (that is, different efficiencies) shown by Fehrer et al. (2006).
O'Callaghan et al. (2008) reported a telomere Q-PCR variation that used oligonucleotides as an external standard to achieve an absolute quantification of TL in kilobase per diploid genome. Unfortunately, the authors did not validate their results, otherwise they would have noticed that the estimated TL of 110-kb telomeric repeats per diploid genome for young adult humans and 80-kb telomeric repeats per diploid genome for old adult humans is equivalent to a mean TL of 1.2 kb for young adults and 0.87 kb for old adults (kb per genome divided by 92 chromosome ends (O'Callaghan et al., 2008)). These values are inconsistent with all human telomere data published to date, even when taking subtelomeric regions measured by TRF analysis into account (Cawthon, 2002; Baird et al., 2006).
Another challenge for telomere Q-PCR that can profoundly affect the results is the choice of the SCG. Many SCGs possess similar pseudo-genes across the genome, yet this is seldom considered. For example, primers used for 36B4 (Cawthon, 2002; Gil and Coetzer, 2004; Fehrer et al., 2006; Brouilette et al., 2007) do not only amplify a region of 36B4, but also an unknown locus on chromosome 2 according to UCSC in silico PCR (http://www.genome.ucsc.edu/cgi-bin/hgPcr) on whole human genome NCBI Build 36.1. Similarly, the GAPDH primers used by Koppelstaetter et al. (2005) amplify not only GAPDH, but also a fragment of ADRA1B on chromosome 5. Although whole genome builds are prone to assembly errors and in silico PCR is not a substitute for PCR, a more careful evaluation of SCG and their primers should be desirable where this is possible, for example, in human and other species in which genomic sequences are available. Some of the genes used as SCG (for example, GAPDH, β2-globin and β-actin) are housekeeping genes commonly used for expression analysis (Bustin, 2000). These housekeeping genes are supposed to have a constant expression level in the cell, but that does not necessarily mean that they are single copy. For example, Hatase et al. (2008) used the 18S ribosomal RNA gene as a SCG to measure TL in loggerhead turtles (Caretta caretta). This gene is a component of rDNA and is known to be present in repeated units on multiple chromosomes, that each can vary in copy number among individuals (Srivastava and Schlessinger, 1991) and thus is completely unsuitable as a single-copy reference gene for telomere Q-PCR.
Despite the alluring simplicity of real-time PCR, conducting these experiments, and especially telomere Q-PCR experiments, with the level of rigour needed to obtain publishable results is not trivial and it is important to fully understand the methodology. For example, as described earlier, the inclusion of an SCG is necessary in telomere Q-PCR to normalize the amount of telomeric sequences to the amount of starting DNA material, as no other DNA quantification method (for example, UV absorbance, fluorescence based approaches) is accurate enough to reliably normalize telomere signal to DNA quantity. Therefore, it is not appropriate to state that the amplification of the SCG, and hence their single-copy state, did not change with age as there was no correlation between the Ct values and age (Criscuolo et al., 2009). A correlation between the Ct of the SCG and age could not possibly be found unless all reactions have exactly the same amount of DNA, and in this case, we would not need the SCG at all to normalize the telomere signal to the DNA amount.
All the above factors can lead to unreliable TL estimates, which sometimes can be identified by the presented amplification efficiencies, coefficient of variation, TL ranges or other inconsistencies (Horn, 2008; Aviv, 2009), but most of the time are not recognizable due to insufficient descriptions of experimental methodology.
Recently, Cawthon (2009) presented a new multiplex telomere Q-PCR that measures SCG and telomere amplification in one reaction tube. Although Cawthon (2009) demonstrated that this method correlates well with TRF in human, it is not clear to what extent it can be applied in ecological studies. Multiplex PCR itself is already a complex method demanding extensive optimization and the new multiplex telomere Q-PCR requires a sequential quantification of SCG and telomere reaction, as the SCG signal is detected well after the telomere reaction started or even finished. It might take a lot of optimization to balance the PCR components between, among other factors, lack of reagents for the SCG reaction, end product inhibition by the telomere product and telomere–primer dimer formation. This method would certainly improve the efficiency of telomere Q-PCR by saving time and consumables, but it remains to be seen whether it can be reproduced accurately in other species and whether it is feasible for ecologists who are not necessarily experts in molecular techniques such as multiplex Q-PCR.
Relationship between TRF and telomere Q-PCR
Another source of methodological error is the conversion of relative TL from T/S values to actual length in kilobase. This is a vital step in order to compare results between studies that use different methods to measure TL. Table 2 shows that the relationship between T/S and the absolute TL (measured by TRF) is not constant among studies. Although most studies do report a high correlation between results obtained by Q-PCR and TRF (Table 2), it is important to recognize that the conversion factors differ. Adoption of a conversion formula from another study, therefore, would result in highly inaccurate estimates for absolute TL in kilobase. Furthermore, some of the published conversion factors are of unknown origin (Thibeault et al., 2006; Njajou et al., 2007) and questionable as they miss one of the two parameters (slope and intercept of the regression) necessary to convert T/S to kilobase.
The differences in conversion factors might also arise from different TRF methods used in these studies (discussed above). Unfortunately, Q-PCR studies that verified their results with TRF tend to describe their TRF method even more briefly than studies focused solely on TRF, and so that it is impossible to distinguish the individual effects of error in the Q-PCR and TRF methods on the regressions. Aviv (2008) already identified this drawback and suggested to set up a panel of standard blinded sets of high-quality DNA by a major organization that could be used by all laboratories to assure the accuracy of their methods. Unfortunately, like many suggestions made for quality control in science (for example, Nolan et al., 2006 for PCR amplification efficiency), this suggestion has thus far gone unheeded.
It is debatable how strong the correlation between TRF and Q-PCR should be. A significant correlation between both methods is often overinterpreted. For example, height and weight or age and length (or even metabolic rate) of most species are significantly correlated, but no one would argue that these are measures of the same variable. It is important to remember that TRF and Q-PCR are supposed to measure the same thing, even if small differences might occur due to subtelomeric sequences in TRF or interstitial telomeric repeats in Q-PCR (Cawthon, 2002; Baird et al., 2006). It is hard to draw a line, but in our opinion R2 values <0.8 indicate that the two methods simply cannot be assumed to measure the same thing.
If done properly, the advantages of telomere Q-PCR over TRF are striking, but authors should be careful not to exaggerate when emphasizing the superiority of Q-PCR. For example, a TRF blot does not require 5–10 μg of DNA, as stated by Criscuolo et al. (2009) or Nakagawa et al. (2004), but only about 0.5–1 μg (for example, Horn et al., 2008), which can be obtained from ∼0.5 μl whole blood in birds and fish (H Thorsten, personal observation). The amount of DNA is not an argument against TRF in small passerine birds and other species with nucleated erythrocytes. However, it can be problematic in mammalian species whose erythrocytes are not nucleated.
Likewise, the disadvantages of Q-PCR should not be trivialized. The primers used to amplify telomeres do also amplify interstitial telomeric repeats, leading to an overestimation of TL depending on the abundance of these repeats. This can be dangerous if the number and size of telomeric repeats varies between individuals in a study. Although Delany et al. (2003) reported low variation of interstitial repeats within inbred lines of chicken (Gallus gallus domesticus), there are differences between inbred lines and especially within non-inbred populations (Delany et al., 2000).
As Aviv (2009) puts it: ‘TRF is indeed labour intensive, costly, requires lots of DNA (for human epidemiology standard, AN) and demands experience and expertise, which explain why this method, considered the gold standard of telomere length measurements, is not now widely used in epidemiology’. The same applies to its use in ecology. However high-throughput and simplicity to perform should always be weighed against reproducibility and reliability when choosing an appropriate method to measure TL.
Conclusion
All the issues we have highlighted in this review contribute to a growing confusion among ecologists and evolutionary biologists, similar to that described by Aviv (2008) for human telomere epidemiology. There is an increasing number of papers published with data generated on TL that report correlations with age and fitness, which on the surface appear quite compelling. In addition, the methodology presented in these papers appears straightforward, and thus numerous laboratories have or are about to start using these methods as supplements to the other suite of molecular tools now widespread in ecological and evolutionary biology laboratories around the globe. However, as we illustrate here, there is significant peril, as well as promise, associated with the use of these methods. In particular, the use of different methods, submethods, and even the erroneous application of these methods and the interpretation of the resultant raw data hamper comparability among studies. Thus, in the current environment, where there is little or no standardization of approaches, it is extremely difficult to find any underpinning relationships between telomere dynamics and parameters of interest to ecologists and evolutionary biologists, such as ageing, life history and fitness through, for example, meta-analysis.
In Supplementary Table S2, we summarize the main characteristics of TRF and Q-PCR, including methodological requirements, advantages and disadvantages, and error sources to assist ecologists and evolutionary biologists in the choice of methods suitable for their study design.
Future directions
The introduction of telomere biology (the study of telomere dynamics within and between species) into ecology and evolutionary studies has opened up a wide field of opportunities. Early promises, namely, telomere ageing, did not hold up to expectations and should give way to the next generation of telomere research. The new focus should be on the factors influencing TL and the subsequent consequences for individual fitness. The main questions we see for future research are:
-
i)
How do environmental conditions and genetic predispositions influence TL? It seems that early life conditions can influence TL change (Hall et al., 2004) and that environmental and/or genetic factors can influence TL (Horn et al., 2008). More investigation in wild populations, but also captive populations under controlled conditions will help us understand the mechanisms leading to the high age-independent variation seen between individuals of a population.
-
ii)
What is the dynamic range of TL and what are the consequences of short telomeres for the individual? Although the time during development and juvenile growth might have a significant influence on TL change later in life, as discussed above, there seems to be a period of rapid TL decline later in life that is closely followed by death, at least in birds. Bize et al. (2009) showed that birds with higher TL attrition rate are more likely to die and Salomons et al. (2009) reported a rapid decline of TL in the last year before disappearance/death. It will be interesting to see whether this rapid decline is induced by a downwards spiral of telomere shortening at the end of life or whether alternative ageing factors induce TL decline in the last life stage in birds and possibly other species.
There is still a large demand for TL data in ecological and evolutionary studies. However, it is important not to repeat the mistakes made in other disciplines (Aviv, 2008), but rather, to design studies to ensure the reliability and transparency of the used methods. The ‘rush to publish’ (Aviv, 2008) has already created a certain degree of confusion in the field of telomere ecology (the study of telomere dynamics in an ecological context), but if we acknowledge the shortcomings of the methods and avoid past mistakes, telomere biology can be an important tool for ecology and help to understand the evolution of fitness trade-offs and ageing in a natural environment.
Although ecologists and evolutionary biologists are often not experts in molecular biology, they nevertheless have an obligation to use the methods correctly and ensure reliability and repeatability of results. We hope this review will contribute to a better understanding of these methods and to a more rigorous application of these tools in the future and urge authors, editors and reviewers to help ensure compatibility among studies to form one big picture of telomere dynamics in wild species.
References
Aubert G, Lansdorp PM (2008). Telomeres and aging. Physiol Rev 88: 557–579.
Aviv A (2006). Telomeres and human somatic fitness. J Gerontol A Biol Sci Med Sci 61: 871–873.
Aviv A (2008). The epidemiology of human telomeres: faults and promises. J Gerontol A Biol Sci Med Sci 63: 979–983.
Aviv A (2009). Commentary: raising the bar on telomere epidemiology. Int J Epidemiol 38: 1735–1736.
Baerlocher GM, Rice K, Vulto I, Lansdorp PM (2007). Longitudinal data on telomere length in leukocytes from newborn baboons support a marked drop in stem cell turnover around 1 year of age. Aging Cell 6: 121–123.
Baird DM (2006). Telomeres. Exp Gerontol 41: 1223–1227.
Baird DM (2008). Telomeres II. Exp Gerontol 43: 15–19.
Baird DM, Britt-Compton B, Rowson J, Amso NN, Gregory L, Kipling D (2006). Telomere instability in the male germline. Hum Mol Genet 15: 45–51.
Bize P, Criscuolo F, Metcalfe NB, Nasir L, Monaghan P (2009). Telomere dynamics rather than age predict life expectancy in the wild. Proc R Soc Lond B Biol Sci 276: 1679–1683.
Bolzan AD, Bianchi MS (2006). Telomeres, interstitial telomeric repeat sequences, and chromosomal aberrations. Mutat Res 612: 189–214.
Brouilette SW, Moore JS, McMahon D, Thompson JR, Ford I, Shepherd J et al. (2007). Telomere length, risk of coronary heart disease, and statin treatment in the West of Scotland Primary Prevention Study: a nested case-control study. Lancet 369: 107–114.
Brummendorf TH, Mak J, Sabo KM, Baerlocher GM, Dietz K, Abkowitz JL et al. (2002). Longitudinal studies of telomere length in feline blood cells: Implications for hematopoietic stem cell turnover in vivo. Exp Hematol 30: 1147–1152.
Bustin SA (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25: 169–193.
Bustin SA, Mueller R (2005). Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin Sci 109: 365–379.
Bustin SA, Nolan T (2004). Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech 15: 155–166.
Callicott RJ, Womack JE (2006). Real-time PCR assay for measurement of mouse telomeres. Comp Med 56: 17–22.
Cawthon RM (2002). Telomere measurement by quantitative PCR. Nucleic Acids Res 30: e47.
Cawthon RM (2009). Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res 37: e21.
Cawthon RM, Smith KR, O'Brien E, Sivatchenko A, Kerber RA (2003). Association between telomere length in blood and mortality in people aged 60 years or older. Lancet 361: 393–395.
Cherkas LF, Aviv A, Valdes AM, Hunkin JL, Gardner JP, Surdulescu GL et al. (2006). The effects of social status on biological aging as measured by white-blood-cell telomere length. Aging Cell 5: 361–365.
Cottliar ASH, Noriega MF, Narbaitz M, Rodriguez A, Slavutsky IR (2006). Association between telomere length and BCL2 gene rearrangements in low- and high-grade non-Hodgkin lymphomas. Cancer Genet Cytogenet 171: 1–8.
Criscuolo F, Bize P, Nasir L, Metcalfe NB, Foote CG, Griffiths K et al. (2009). Real-time quantitative PCR assay for measurement of avian telomeres. J Avian Biol 40: 342–347.
de Lange T (2002). Protection of mammalian telomeres. Oncogene 21: 532–540.
de Lange T (2005). Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110.
Delany ME, Daniels LM, Swanberg SE, Taylor HA (2003). Telomeres in the chicken: genome stability and chromosome ends. Poult Sci 82: 917–926.
Delany ME, Krupkin AB, Miller MM (2000). Organization of telomere sequences in birds: evidence for arrays of extreme length and for in vivo shortening. Cytogenet Cell Genet 90: 139–145.
Demerath EW, Cameron N, Gillman MW, Towne B, Siervogel RM (2004). Telomeres and telomerase in the fetal origins of cardiovascular disease: a review. Hum Biol 76: 127–146.
Demissie S, Levy D, Benjamin EJ, Cupples LA, Gardner JP, Herbert A et al. (2006). Insulin resistance, oxidative stress, hypertension, and leukocyte telomere length in men from the Framingham Heart Study. Aging Cell 5: 325–330.
Epel ES, Blackburn EH, Lin J, Dhabhar FS, Adler NE, Morrow JD et al. (2004). Accelerated telomere shortening in response to life stress. Proc Natl Acad Sci USA 101: 17312–17315.
Epel ES, Lin J, Wilhelm FH, Wolkowitz OM, Cawthon R, Adler NE et al. (2006). Cell aging in relation to stress arousal and cardiovascular disease risk factors. Psychoneuroendocrinology 31: 277–287.
Fehrer C, Voglauer R, Wieser M, Pfister G, Brunauer R, Cioca D et al. (2006). Techniques in gerontology: cell lines as standards for telomere length and telomerase activity assessment. Exp Gerontol 41: 648–651.
Frenck Jr RW, Blackburn EH, Shannon KM (1998). The rate of telomere sequence loss in human leukocytes varies with age. Proc Natl Acad Sci USA 95: 5607–5610.
Gil ME, Coetzer TL (2004). Real-time quantitative PCR of telomere length. Mol Biotechnol 27: 169–172.
Grabowski P, Hultdin M, Karlsson K, Tobin G, Aleskog A, Thunberg U et al. (2005). Telomere length as a prognostic parameter in chronic lymphocytic leukemia with special reference to V-H gene mutation status. Blood 105: 4807–4812.
Grant JD, Broccoli D, Muquit M, Manion FJ, Tisdall J, Ochs MF (2001). Telometric: a tool providing simplified, reproducible measurements of telomeric DNA from constant field agarose gels. Biotechniques 31: 1314–1318.
Hall ME, Nasir L, Daunt F, Gault EA, Croxall JP, Wanless S et al. (2004). Telomere loss in relation to age and early environment in long-lived birds. Proc R Soc Lond B Biol Sci 271: 1571–1576.
Harley CB, Futcher AB, Greider CW (1990). Telomeres shorten during ageing of human fibroblasts. Nature 345: 458–460.
Harris SE, Deary IJ, Maclntyre A, Lamb KJ, Radhakrishnan K, Starr JM et al. (2006). The association between telomere length, physical health, cognitive ageing, and mortality in non-demented older people. Neurosci Lett 406: 260–264.
Hartmann N, Reichwald K, Lechel A, Graf M, Kirschner J, Dorn A et al. (2009). Telomeres shorten while Tert expression increases during ageing of the short-lived fish Nothobranchius furzeri. Mech Ageing Dev 130: 290–296.
Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC (1990). Telomere reduction in human colorectal-carcinoma and with aging. Nature 346: 866–868.
Hatakeyama H, Nakamura KI, Izumiyama-Shimomura N, Ishii A, Tsuchida S, Takubo K et al. (2008). The teleost Oryzias latipes shows telomere shortening with age despite considerable telomerase activity throughout life. Mech Ageing Dev 129: 550–557.
Hatase H, Sudo R, Watanabe KK, Kasugai T, Saito T, Okamoto H et al. (2008). Shorter telomere length with age in the loggerhead turtle: a new hope for live sea turtle age estimation. Genes Genet Syst 83: 423–426.
Haussmann MF, Mauck RA (2008). New strategies for telomere-based age estimation. Mol Ecol Resour 8: 264–274.
Haussmann MF, Vleck CM (2002). Telomere length provides a new technique for aging animals. Oecologia 130: 325–328.
Haussmann MF, Vleck CM, Nisbet ICT (2003a). Calibrating the telomere clock in common terns, Sterna hirundo. Exp Gerontol 38: 787–789.
Haussmann MF, Winkler DW, O'Reilly KM, Huntington CE, Nisbet ICT, Vleck CM (2003b). Telomeres shorten more slowly in long-lived birds and mammals than in short-lived ones. Proc R Soc Lond B Biol Sci 270: 1387–1392.
Haussmann MF, Winkler DW, Vleck CM (2005). Longer telomeres associated with higher survival in birds. Biol Lett 1: 212–214.
Hewakapuge S, van Oorschot RA, Lewandowski P, Baindur-Hudson S (2008). Investigation of telomere lengths measurement by quantitative real-time PCR to predict age. Leg Med 10: 236–242.
Horn T (2008). Comments on quantitative real-time pcr for measurement of telomere length. Cancer Invest 26: 867.
Horn T (2009). Telomere length of Kakapo and other New Zealand birds—assessment of methods and applications. PhD Thesis, University of Canterbury, Christchurch, New Zealand.
Horn T, Gemmell NJ, Robertson BC, Bridges CR (2008). Telomere length change in European sea bass (Dicentrarchus labrax). Aust J Zool 56: 207–210.
Juola FA, Haussmann MF, Dearborn DC, Vleck CM (2006). Telomere shortening in a long-lived marine bird: cross-sectional analysis and test of an aging tool. Auk 123: 775–783.
Kirk KE, Harmon BP, Reichardt IK, Sedat JW, Blackburn EH (1997). Block in anaphase chromosome separation caused by a telomerase template mutation. Science 275: 1478–1481.
Koppelstaetter C, Jennings P, Hochegger K, Perco P, Ischia R, Karkoszka H et al. (2005). Effect of tissue fixatives on telomere length determination by quantitative PCR. Mech Ageing Dev 126: 1331–1333.
Londono-Vallejo JA, DerSarkissian H, Cases L, Thomas G (2001). Differences in telomere length between homologous chromosomes in humans. Nucleic Acids Res 29: 3164–3171.
Martin-Ruiz C, Saretzki G, Petrie J, Ladhoff J, Jeyapalan J, Wei WY et al. (2004). Stochastic variation in telomere shortening rate causes heterogeneity of human fibroblast replicative life span. J Biol Chem 279: 17826–17833.
Mefford HC, Trask BJ (2002). The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet 3: 91–102.
Meyne J, Ratliff RL, Moyzis RK (1989). Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci USA 86: 7049–7053.
Monaghan P, Haussmann MF (2006). Do telomere dynamics link lifestyle and lifespan? Trends Ecol Evol 21: 47–53.
Nakagawa S, Gemmell NJ, Burke T (2004). Measuring vertebrate telomeres: applications and limitations. Mol Ecol 13: 2523–2533.
Njajou OT, Cawthon RM, Damcott CM, Wu SH, Ott S, Garant MJ et al. (2007). Telomere length is paternally inherited and is associated with parental lifespan. Proc Natl Acad Sci USA 104: 12135–12139.
Nolan T, Hands RE, Bustin SA (2006). Quantification of mRNA using real-time RT-PCR. Nat Protoc 1: 1559–1582.
Nordfjall K, Larefalk A, Lindgren P, Holmberg D, Roos G (2005). Telomere length and heredity: Indications of paternal inheritance. Proc Natl Acad Sci USA 102: 16374–16378.
Nordgard O, Kvaloy JT, Farmen RK, Heikkila R (2006). Error propagation in relative real-time reverse transcription polymerase chain reaction quantification models: The balance between accuracy and precision. Anal Biochem 356: 182–193.
O'Callaghan N, Dhillon V, Thomas P, Fenech M (2008). A quantitative real-time PCR method for absolute telomere length. Biotechniques 44: 807–809.
Pauliny A, Wagner RH, Augustin J, Szep T, Blomqvist D (2006). Age-independent telomere length predicts fitness in two bird species. Mol Ecol 15: 1681–1687.
Pfaffl MW (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45.
Rebrikov DV, Trofimov DY (2006). Real-time PCR: a review of approaches to data analysis. Appl Biochem Microbiol 42: 455–463.
Rufer N, Brummendorf TH, Kolvraa S, Bischoff C, Christensen K, Wadsworth L et al. (1999). Telomere fluorescence measurements in granulocytes and T lymphocyte subsets point to a high turnover of hematopoietic stem cells and memory T cells in early childhood. J Exp Med 190: 157–167.
Salomons HM, Mulder GA, van de Zande L, Haussmann MF, Linskens MHK, Verhulst S (2009). Telomere shortening and survival in free-living corvids. Proc R Soc Lond B Biol Sci 276: 3157–3165.
Sambrook J, Russell D (2001). W. 01. Gel electrophoresis. In: Molecular Cloning. A Laboratory Manual. Cold Spring Harbour Laboratory: New York.
Scott NM, Haussmann MF, Elsey RM, Trosclair PL, Vleck CM (2006). Telomere length shortens with body length in Alligator mississippiensis. Southeast Nat 5: 685–692.
Srivastava AK, Schlessinger D (1991). Structure and organization of ribosomal DNA. Biochimie 73: 631–638.
Stratagene (2004). Introduction to Quantitative PCR, Methods and Application Guide. IN #70200-00/Revision #084001.
Thibeault SL, Glade TS, Li WH (2006). Comparison of telomere length of vocal folds with different tissues: a physiological measurement of vocal senescence. J Voice 20: 165–170.
Traut W, Szczepanowski M, Vitkova M, Opitz C, Marec F, Zrzavy J (2007). The telomere repeat motif of basal metazoa. Chromosome Res 15: 371–382.
Valdes AM, Andrew T, Gardner JP, Kimura M, Oelsner E, Cherkas LF et al. (2005). Obesity, cigarette smoking, and telomere length in women. Lancet 366: 662–664.
Von Zglinicki T (2002). Oxidative stress shortens telomeres. Trends Biochem Sci 27: 339–344.
Wilson IG (1997). Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol 63: 3741–3751.
Wright WE, Shay JW (2005). Telomere biology in aging and cancer. J Am Geriatr Soc 53: S292–S294.
Zhang Y, Shen J, Ming W, Lee YPH, Santella RM (2007). Telomere length in hepatocellular carcinoma and paired adjacent non-tumor tissues by quantitative PCR. Cancer Invest 25: 668–677.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Horn, T., Robertson, B. & Gemmell, N. The use of telomere length in ecology and evolutionary biology. Heredity 105, 497–506 (2010). https://doi.org/10.1038/hdy.2010.113
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2010.113
Keywords
This article is cited by
-
No evidence of physiological declines with age in an extremely long-lived fish
Scientific Reports (2021)
-
Shorter telomeres precede population extinction in wild lizards
Scientific Reports (2017)
-
Different effects of accelerated development and enhanced growth on oxidative stress and telomere shortening in amphibian larvae
Scientific Reports (2017)
-
Environmental, not individual, factors drive markers of biological aging in black bears
Evolutionary Ecology (2017)
-
Repeatability of a terminal restriction fragment telomere length assay on red drum fin tissue
Conservation Genetics Resources (2017)