Abstract
The living fossil Triops cancriformis comprises bisexual (either gonochoric or hermaphroditic) and unisexual populations. Genetic surveys have recently revealed a general trend of low differentiation of 12S and 16S mitochondrial genes. We, therefore, surveyed further mitochondrial (COI gene and control region) and nuclear markers (dinucleotide microsatellites) to assess the genetic variability and to establish any relationship with the different reproductive modes found in European populations. The mitochondrial analyses confirmed the pattern of low variability. Hence, the low mitochondrial genetic variability appears as a common feature of the genus Triops. The microsatellite analysis found that Italian populations are monomorphic or exhibit little polymorphism, while other European samples display a higher degree of polymorphism and private alleles. Spanish, Austrian and Italian populations show patterns of Hardy–Weinberg disequilibrium that could be explained by the mode of reproduction, or by a higher frequency of null alleles in these populations. The low diversity and differentiation among Italian populations lead us to question the Monopolization Hypothesis. One microsatellite locus appears to be sex-linked, with heterozygotes detected only in males and hermaphrodites.
Similar content being viewed by others
Introduction
The living fossil Triops cancriformis (Bosc, 1801) (Crustacea, Branchiopoda, Notostraca) lives in Eurasian and North African astatic waters with bisexual and unisexual populations. While the latter comprises parthenogenetic females with an all-female progeny (thelytoky), the former can be either gonochoric, with distinct female and male individuals, or hermaphroditic with individuals producing both male and female gametes (Trusheim, 1938; Longhurst, 1955; Wingstrand, 1978; Zaffagnini and Trentini, 1980; Fryer, 1985; Eder et al., 1996; Engelmann et al., 1997; Scanabissi and Mondini, 2002; Scanabissi et al., 2005). The sexuality of a population is difficult to determine on morphological grounds, the only diagnostic characters being the modification of the eleventh pair of trunk appendages in both sexes and the presence/absence of eggs (Mathias, 1937; Fryer, 1988; Engelmann et al., 1996). These characters are often misleading (hermaphrodites with eggs can be mistakenly recognized as females) or inapplicable (young individuals). Moreover, the diffusion of resistant eggs by means of wind or birds (Figuerola et al., 2005) promotes migration fluxes, so that individuals deriving from populations with different sexuality may occur in the same site. A remarkable example is the finding of functional males (Scanabissi et al., 2005) in a hermaphroditic Austrian population (sensu Wingstrand, 1978). This could be due to the introduction of new resistant eggs, but the co-occurrence of males and hermaphrodites suggests that this T. cancriformis might be considered androdioecious (a condition in which hermaphrodites and functional males coexist; Pannell, 2002; Weeks et al., 2006).
Besides its sexuality, the taxonomy of T. cancriformis populations has always been controversial (Ghigi, 1921, 1924; Colosi, 1923; Gurney, 1923; Gauthier, 1934) owing to the high variability of individual morphological characters (Longhurst, 1955; Alonso, 1985). In his monograph on Notostraca, Longhurst (1955) finally acknowledged the presence of three subspecies, T. cancriformis cancriformis, T. cancriformis mauritanicus Ghigi, 1921 and T. cancriformis simplex (Ghigi, 1921). Among the morphological characters used by Longhurst, Alonso (1985) recognized the presence/absence of spines in the carapace carina as the only useful taxonomic character, the alternatives being exceedingly variable, with differences even between the left and right sides of the same individual.
A recent molecular analysis on 12S and 16S mitochondrial markers based on a wide taxon sampling (Korn et al., 2006) indicates that T. cancriformis is divided into two distinct lineages. One lineage comprises European T. cancriformis cancriformis populations and individuals from Northern Spain that had been classified as T. cancriformis simplex in the most recent literature (Alonso, 1985, 1996; Boix et al., 2002). The second lineage comprises other Iberian and the North African populations of T. cancriformis simplex and T. cancriformis mauritanicus populations. The authors, therefore, propose that the two lineages are recognized as distinct species, T. cancriformis and T. mauritanicus, respectively. Even if the level of divergence emerging from the analyses appears low, for our present purposes we accept the new terminology, which provides some clarity.
While the strictly gonochoric T. mauritanicus shows high substructuring and may include at least five subspecies, the amphigonic and parthenogenetic populations of T. cancriformis are more uniform (Korn et al., 2006). This difference had previously been observed in a survey within T. cancriformis populations (sensu Korn et al., 2006), where a very low level of variability was found for the 12S and 16S genes (Mantovani et al., 2004). The same paper demonstrated also that T. cancriformis was significantly differentiated from a selection of congeneric taxa, thus supporting the hypothesis that this species should be ascribed to a separated genus (Linder, 1952).
The present study was undertaken to investigate whether the low level of genetic variability reported for T. cancriformis was the consequence of the particular choice of molecular markers (12S and 16S) or some property of the mitochondrial genome. We, therefore, use different mitochondrial genes (the cytochrome oxidase I gene and the mitochondrial control region) and some previously identified dinucleotide microsatellite loci from the nuclear genome (Cesari et al., 2004). The mitochondrial markers have proved very useful in previous crustacean genetic studies (Remigio and Hebert, 2000; Chu et al., 2003), while microsatellites are known for their high polymorphism and usually provide a powerful tool for population genetic studies.
Given the reproductive variability within T. cancriformis, in this paper we analyze gonochoric, hermaphroditic and parthenogenetic T. cancriformis populations collected in Spain, Austria and Italy, respectively. We want to establish whether the genetic variation is higher in bisexually reproducing populations (either gonochoric or hermaphroditic) than in the parthenogenetic ones and if both mitochondrial and nuclear compartments show the same trend.
Materials and methods
Animals
The work was carried out on seven populations. Most localities were sampled once, with the exception of Grosseto and Espolla, which were examined in two different years; a total number of nine samples were therefore considered (Table 1; Figure 1).
In each sample, two individuals were analyzed for all four mitochondrial genes (Table 1). Ten additional specimens were analyzed for the 16S gene in all samples but Palermo.
From 17 to 22 individuals were genotyped at the five microsatellite loci (MSL) in six samples; for Marchegg and Oristano populations we considered previous data (Cesari et al., 2004), while Palermo was not surveyed because too few specimens were available.
Total DNA was extracted from single individuals, following cetyltrimethylammonium bromide (CTAB) (Doyle and Doyle, 1987) or phenol/chloroform (Sambrook et al., 1989) protocols.
Mitochondrial analyses
PCR amplification was performed in 50 μl reactions using the Invitrogen PCR kit with recombinant Taq DNA polymerase. A total of 35 cycles were scheduled as follows: denaturation at 94 °C for 30 s, annealing at 48 °C for 30 s, extension at 72 °C for 30 s. The amplified products were purified with the Wizard PCR cleaning (Promega, Madison, WY, USA) kit and both strands were sequenced in an ABI PRISM 310 Genetic Analyzer (Applera, Norwalk, CT, USA). The primers for PCR amplification and sequencing are reported in Table 2. Primers for the control region were specifically designed on the T. cancriformis complete mitochondrial sequence (MCR; GenBank accession number NC_004465). Alignments were performed using the Clustal algorithm of the Sequence Navigator program (version 1.0.1, Applera; multiple alignment parameters: Gap Open Penalty=10, Gap Extend Penalty=10). The nucleotide sequences of the newly analyzed specimens have been submitted to GenBank (accession numbers DQ369307-8, 12S; DQ369309, DQ664195 and EF190477-81, 16S; DQ369312-7, DQ664196, COI; AY764144-6, DQ369310-1 and EF190476, MCR). Homologous sequences were drawn from the complete mitochondrial sequences of a Japanese T. cancriformis (accession number NC_004465) and of Triops longicaudatus LeConte, 1846 (accession NC_006079); the latter was used as outgroup. The observed number of substitutions was determined using MEGA version 3.1 (Kumar et al., 2004), while Maximum Parsimony (MP) and Maximum Likelihood (ML) dendrograms were computed using PAUP* 4.0b10 (Swofford, 2001); bootstrap values were obtained after 2000 and 200 replicates, respectively. MP analysis was performed with tree bisection-reconnection branch-swapping algorithm and stepwise addition of taxa with 100 random replicates; gaps were considered as fifth state. For ML analysis, Modeltest (version 3.06; Posada and Crandall, 1998) was run to determine the best substitution model (TVM+G), with the evaluation of base frequencies, rate matrix, proportion of invariable sites and value of γ-shaped parameter (0.2113). A parsimony network was determined for the 16S haplotypes coming from 12 individuals for each population, but Palermo, by applying the method described by Templeton et al. (1992) as implemented in TCS 1.21 (Clement et al., 2000), with gaps considered as fifth state.
Microsatellites analyses
Populations were genotyped at five dinucleotide microsatellite loci (tcAC-8p1, tcAC-9p1, tcAC-10p1, tcAC-10p2 and tcAC-14p1) following the protocols described by Cesari et al. (2004). Observed and expected heterozygosities, allelic frequencies and estimated number of migrants (Nm, following Wright, 1969) were computed using Genetix 4.05 (Belkhir et al., 2004). Hardy–Weinberg equilibrium (HWE), linkage disequilibrium test, the relationship between population differentiation and geographical distance, genic and genotypic differentiation were calculated using Genepop 1.2 (Raymond and Rousset, 1995). Allelic richness and F-statistics were computed using FSTAT 2.9.3 (Goudet, 2001). F-statistics were also obtained from previously analyzed Italian and Austrian samples (Cesari et al., 2004). Population differentiation based on microsatellite frequencies and the number of migrants (M=2 Nm, where Nm=((1−FST)/(2 × FST))) were calculated with Arlequin 3.0b (Excoffier et al., 2005). Given that polymorphism at annealing sites of the MSL primers can prevent the amplification of a particular allele, therefore resulting in heterozygote deficiencies, null allele frequencies were estimated as described by Chakraborty et al. (1992); Brookfield (1996); Van Oosterhout et al. (2004), with Bonferroni corrections, using Microchecker 2.2.1 (Van Oosterhout et al. 2004).
Results
Mitochondrial analysis
mtDNA diversity
Overall, 1816–1823 base pairs were sequenced in each individual (12S gene: 347 bp; 16S gene: 503–509 bp; COI gene: 595 bp; MCR: 371–372 bp), and 14 mitotypes (that is, combined mitochondrial haplotypes) are presented in Table 1. All populations had private mitotypes. A single mitotype characterized Lecce and Espolla 2006. In the Grosseto pond, differences were found between samples obtained in consecutive years (2002–2003), but not between specimens sampled the same year. In each of the other populations, the two individuals showed mitotypes differing for one/two point mutations or for one indel, with the exception of the two Marchegg individuals, which differed for 29 substitutions.
The Marchegg mitotype A was the most distinct from any other (29–33 substitutions), while mitotype B differed for only 1–15 substitution(s). The comparison between Italian mitotypes showed at most four substitutions, with the exception of the Sicilian and Apulian populations, which were distinguished by 13–17 substitutions. On the whole, the four Spanish specimens were more similar to the Sardinian sample (two indels or 2–3 substitutions).
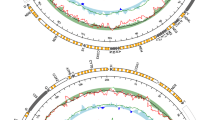
MP and ML dendrograms (Figure 2) differ in deep branching topology. In the MP tree (single trees’ island with four equally parsimonious trees; Figure 2a), the Austrian mitotype A is basally located and two further highly supported clusters can be recognized. On the other hand, the ML dendrogram (Figure 2b) is mainly polytomic, with the Austrian mitotype A occurring in the only supported cluster with Apulian and Sicilian sequences. The Spanish and Sardinian mitotypes appear more related to each other in the MP analysis, while their cluster collapses in the ML analysis.
(a) Maximum Parsimony (MP) phylogram (consistency index: 0.974; retention index: 0.839; tree length: 349) computed on the combined mitochondrial dataset (12S, 16S, MCR and COI). Values above the branches indicate mutational steps, while those under the branches show bootstrap values. (b) Maximum Likelihood (−lnL: 3824.69) phylogram obtained from combined analyses of the four mitochondrial genes. Values below the branches represent bootstrap percentages. Color codes indicating each locality are shown in Figure 1 (the black dot with the star represents the Japanese, GenBank drawn mitotype).
16S gene haplotype differentiation
The mitochondrial analysis was performed on 10 more individuals in all populations but Palermo (total number of analyzed specimens=96) for the 16S gene, which was chosen as it proved to be the most variable in the first part of the analysis (Table 1). The newly sequenced specimens revealed five new haplotypes, four in Espolla (i, j, k, l) and one in Marchegg (m), differing respectively for one or two substitutions with respect to the most common haplotype b.
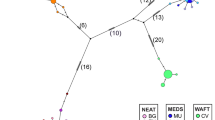
The network analysis produced two different lineages (Figure 3), one comprising only haplotype a, which was found in most Marchegg individuals and in one Oristano specimen. The other lineage included the remaining sequences with haplotype b as the most frequent: it is found in the great majority of Ferrara, Grosseto, Oristano and Espolla individuals, and in one Austrian specimen. Lecce is the only sample presenting a single haplotype e.
16S haplotype parsimony network. Lines represent a single mutational event or an indel regardless of their length, small open dot indicates missing/ideal haplotype, while circles represent haplotypes, with size proportional to the frequency of occurrence. Letters denoting different haplotypes as shown in Table 1, with the addition of haplotypes i–m (see text for details). Color codes indicating each locality are shown in Figure 1 (the black dot with the star represents the Japanese, GenBank drawn mitotype).
Microsatellite analysis
Locus tcAC10-p2 is monomorphic in all six newly genotyped samples. The other loci are all polymorphic in the bisexual Espolla samples, with the presence of private alleles (Table 3, Figure 4). Italian samples show a decidedly low variability; in particular, Grosseto 2003 and Lecce are homozygous at all loci, whereas only one (tcAC10-p1) or two (tcAC9-p1, tcAC10-p1) MSL are polymorphic in the Ferrara and Grosseto 2002 populations, respectively. Null alleles may be present in the Ferrara (locus tcAC10-p1), Grosseto 2002 (locus tcAC9-p1) and Espolla 2004 (loci tcAC8-p1 and tcAC9-p1) samples (Table 3). Moreover, the re-analysis of the data presented by Cesari et al. (2004) with recently developed algorithms (Chakraborty et al., 1992; Van Oosterhout et al., 2004) revealed a probable presence of null alleles in all loci (but tcAC8-p1) in the Marchegg population.
No evidence of significant linkage was found among the five loci (P>0.05). Ferrara and Grosseto 2002 samples deviate significantly from HWE (P<0.05 and P<0.001, respectively, Table 3). The HW disequilibrium over all loci is reflected in the highly significant FIS and FIT values (0.451 and 0.836, respectively; P<0.001). A significant value of FST (0.702; P<0.001) points to a substantial genetic differentiation over all loci.
The pairwise FST values (Table 4) highlight a genetic structuring both between the hermaphroditic Austrian and gonochoric Spanish samples and between them and the Italian populations. Among the latter, the FST values point to a high differentiation of Lecce from the other Italian populations. A significant value is also obtained in the comparison between Grosseto 2003 and Oristano. Genic, genotypic and microsatellite frequency differentiation completely confirm this pattern, even though in the latter analysis the comparison between Grosseto 2003 and Oristano samples is not significant (data available from the authors).
The number of migrants was estimated using two different algorithms (Table 4). Both analyses agree in showing a very low number of migrants between Spanish and Austrian samples and between these populations and the Italian ones. Values indicative of dispersal have been scored among Italian samples with the exception of Lecce, with the highest score found between Ferrara and Grosseto 2003 in the Wright's model. Population differentiation appears correlated with the increasing geographical distance between populations (R2=0.699, P<0.05).
In the Espolla samples, the MSL genotypes were also considered in the light of the specimens’ sex. In the 2004 sample a peculiar sex-linked pattern of variability was found at the tcAC8-p1 locus; the 10 analyzed females (Table 3) had the same homozygous genotype (150/150), while the 10 male specimens had either homozygous or heterozygous genotypes (144/144, 5 males; 144/150, 4 males; 150/150, 1 male). Seventeen additional females of the Espolla 2004 sample were analyzed at this locus, and all of them exhibited the 150/150 genotype. A comparable situation (homozygous females and homozygous or heterozygous males) was found in the 2006 Espolla sample. Furthermore, all 92 Italian females shared the 150/150 genotype (present data; Cesari et al., 2004), while the hermaphroditic Austrian sample had homozygous and heterozygous genotypes (148/148, 4 individuals; 148/150, 6 individuals; 150/150, 2 individuals; Cesari et al., 2004). On the whole, alleles 144 or 148 occur only in males or hermaphrodites in either homozygous (144/144; 148/148) or heterozygous (144/150; 148/150) condition, while the females have only the 150/150 genotype.
Discussion
The mitochondrial analyses confirmed the low variability among T. cancriformis specimens (Mantovani et al., 2004). While this pattern might have been anticipated for a protein-coding gene such as COI, it was absolutely unexpected for the mitochondrial control region, a marker widely used for population analysis (Chu et al., 2003; Kang et al., 2005; Vianna et al., 2006). Also, the widening of the analysis to a higher number of individuals per population for the 16S gene confirms the low differentiation. The most differentiated mitotype (A) shows a pairwise sequence difference percentage ranging from 1.64 to 1.86%: this datum is in line with that found by Murugan et al. (2002) in American T. longicaudatus (12S rDNA, up to 1.6%), but it is lower with respect to sequence divergence found within taxa of the other Notostraca genus, Lepidurus (12S rDNA, up to 3.4%; King and Hanner, 1998).
The mutation rate of mitochondrial DNA is usually very high, and with enough sequence length the error in reconstructing the true species’ genealogy should be small (even if the definition of ‘enough sequence length’ can be problematic; Ballard and Whitlock, 2004). If the time scale for divergence is too small, the number of mutational differences in the populations would be too little, thus preventing accurate reconstruction of the gene trees, even with complete mitochondrial DNA sequence data (Ballard and Whitlock, 2004). Even if T. cancriformis lineage is 200 Myr old (Fryer, 1985), the origin of European populations analyzed here would be expected to date back to 1.08–0.26 Myr ago (Korn et al., 2006). The very recent divergence from the common ancestor could be the cause of the observed low variability. The 16S analysis revealed the presence of a widespread haplotype b, found in 64 of 96 individuals and in 5 of 7 populations. Therefore, the low variability of present mitochondrial markers appears strictly linked to Triops genome. In light of the low diversity, our investigation suggests at most an affinity between Southern Italy populations (Lecce and Palermo) and between Oristano and Espolla mitotypes.
No relationship between the number of haplotypes and the reproductive strategy seems to exist; three mitotypes out of four analyzed individuals can be scored in the bisexual populations of Espolla and eight mitotypes out of 10 analyzed individuals in the parthenogenetic Italian populations.
On the whole, mitochondrial analysis fails to reveal either patterns of population structure or any relationship between variability and reproductive biology.
On the other hand, in microsatellite analyses, FST values point to a clear differentiation between Italian, Austrian and Spanish populations, possibly consistent with a model of isolation by geographic distance. Furthermore, variability levels are quite different among the samples, that is, Italian populations are monomorphic (Grosseto 2003 and Lecce) or show little polymorphism and deviate from HWE because of a significant heterozygote deficiency (Grosseto 2002 and Ferrara). The overall low polymorphism level found in Italian samples is comparable with that previously scored in the Sardinian population (Cesari et al., 2004). Conversely, the gonochoric Spanish samples and the previously analyzed hermaphroditic Austrian population display a higher degree of polymorphism and private alleles.
The clear-cut difference emerging between Spanish, Austrian and Italian samples may be explained by different reasons. First, HW disequilibrium could be linked to null allele presence; however, it should be remembered that the methods to estimate the presence of null alleles assume that we are dealing with gonochoric populations; population subdivision and/or local breeding structure (Brookfield, 1996) are not taken into account. HWE deviations may, therefore, be related to the presence of null alleles in the Spanish gonochoric population, while in Italian and Austrian samples, HWE deviations may be explained by their parthenogenetic (Scanabissi and Mondini, 2002) and hermaphroditic (Wingstrand, 1978) conditions, respectively. On the whole, the differences emerging in variability among Spanish, Austrian and Italian samples appear to be associated with their sexual condition. The presence of a male in the Lecce sample represents the first and so far unique finding of a T. cancriformis male in Italy, and may explain the differentiation of this population from the other Italian ones. Obviously, the reproductive role of this male needs to be clarified, especially in the light of the lack of variability found.
Our analyses reveal a loss of variability in Grosseto in two consecutive years (2002–2003), a pattern also observed with the mitochondrial markers (Table 1). This could be explained by new resistant eggs introduced by migrating birds, although the genetic evidence does not confirm such migration. The new eggs could have hatched in 2002, but then individuals may have been selected against. De Meester et al. (2002) has suggested that strong founder events shape population structure in many aquatic organisms (the Monopolization Hypothesis), with the presence of egg banks creating a powerful buffer against the impact of new migrants. However, the differentiation among Italian samples does not support the theory that the capacity of resource monopolization by obligate parthenogens is low (De Meester et al., 2002). Furthermore, in rotifers with resting egg banks (Gómez and Carvalho, 2000) and in T. longicaudatus (Scott and Grigarick, 1979), there are different types of resistant eggs, which need different conditions of flooding, soil depth and temperature variation to trigger hatching. It is, therefore, possible that the genetic variability detected in the Grosseto 2002 sample was absent in the Grosseto 2003 sample because most eggs did not hatch.
Interestingly, the Espolla sample exhibits all polymorphic loci in HWE, with the notable exception of tcAC8-p1 in the 2004 sample (Table 3). Again, this peculiar situation could be caused by null alleles, but an alternative explanation may be that tcAC8-p1 is a sex-linked locus. In fact, despite the high number of analyzed individuals, all Italian and Spanish females have the same homozygous genotype (150/150), while the Spanish males and the hermaphroditic Austrian specimens also have heterozygous genotypes. It could, therefore, be argued that this locus is sex-linked and that female may be the heterogametic sex. The variability found in males and hermaphrodites may be further explained if the mutations rate is higher in males, possibly owing to a larger number of germ cell divisions (Ellegren, 2000). Obviously, the absence of a linkage map and the inability to identify sex chromosomes due to their very small size (Marescalchi et al., 2005) constitute a consistent drawback. The presence of diagnostic genotypes could, nevertheless, prove very useful for future studies on reproductive biology, and also for conservation purposes.
On the whole, the genetic variation that we detected was low, even in comparison to other parthenogenetic crustacean taxa (Pálsson, 2000; Pfrender et al., 2000). Further studies are, therefore, required to understand the apparently frozen genetic structure of this living fossil.
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
Alonso M (1985). A survey of the Spanish Euphyllopoda. Misc Zool 9: 179–208.
Alonso M (1996). Crustacea, Branchiopoda. In: Ramos MA et al. (eds). Fauna Ibérica, vol. 7, Museo Nacional de Ciencias Naturales, CSIC: Madrid, 486 pp.
Ballard JWO, Whitlock MC (2004). The incomplete natural history of mitochondria. Mol Ecol 13: 729–744.
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004). GENETIX 4.05, logiciel sous Windows™ pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier (France). (code available at http://www.univ-montp2.fr/~genetix/genetix/genetix.htm).
Boix D, Sala J, Moreno-Amich R (2002). Population dynamics of Triops cancriformis (Crustacea: Branchiopoda: Notostraca) of the Espolla temporary pond in the northeastern Iberian peninsula. Hydrobiologia 486: 175–183.
Brookfield JFY (1996). A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol 5: 453–455.
Cesari M, Mularoni L, Scanabissi F, Mantovani B (2004). Characterization of dinucleotide microsatellite loci in the living fossil tadpole shrimp Triops cancriformis (Crustacea Branchiopoda Notostraca). Mol Ecol Notes 4: 733–735.
Chakraborty R, De Andrade M, Daiger SP, Budowle B (1992). Apparent heterozygote deficiencies observed in DNA typing data and their implications in forensic applications. Ann Hum Genet 56: 45–47.
Chu K, Li CP, Tam YK, Lavery S (2003). Application of the mitochondrial control region in population genetic studies of the shrimp Penaeus. Mol Ecol Notes 3: 120–122.
Clement M, Posada D, Crandall K (2000). TCS: a computer program to estimate gene genealogies. Mol Ecol 9: 1657–1660.
Colosi G (1923). Note sopra alcuni Eufillopodi. Atti Soc Ital Sci Nat 62: 75–80.
De Meester L, Gómez A, Okamura B, Schwenk K (2002). The Monopolization Hypotesis and the dispersal-gene flow paradox in aquatic organism. Acta Oecol 23: 121–135.
Doyle JJ, Doyle JL (1987). A rapid DNA isolation method for small quantities of fresh tissues. Phytochemical Bulletin 19: 11–15.
Eder E, Hödl W, Milasowszky N (1996). Die Groß-Branchiopoden des Seewinkels. Stapfia 42: 93–101.
Ellegren H (2000). Evolution of the avian sex chromosomes and their role in sex determination. Trends Ecol Evol 15: 188–192.
Engelmann M, Hahn T, Hoheisel G (1997). Ultrastructural characterization of the gonads of Triops cancriformis (Crustacea, Notostraca) from populations containing both females and males: no evidence for hermaphroditic reproduction. Zoomorphology 117: 175–180.
Engelmann M, Hoheisel G, Hahn T, Joost W, Vieweg J, Naumann W (1996). Populationen von Triops cancriformis (Bosc) (Notostraca) in Deutschland Nördlich 50°N sind nicht klonal und höchstens fakultativ hermaphroditisch. Crustaceana (Leiden) 69: 755–768.
Excoffier L, Laval G, Schneider S (2005). Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1: 47–50.
Figuerola J, Green AJ, Michot TC (2005). Invertebrate eggs can fly: evidence of waterfowl-mediated gene flow in aquatic invertebrates. Am Nat 165: 274–280.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994). DNA primers for amplification of mitochondrial cytochrome oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech 3: 294–299.
Fryer G (1985). Structure and habits of living branchiopod crustaceans and their bearing on the interpretation of fossil forms. T Roy Soc Edin 76: 103–113.
Fryer G (1988). Studies on the functional morphology and biology of the Notostraca (Crustacea: Branchiopoda). Philos Trans R Soc Lond B Biol Sci 321: 27–124.
Gauthier H (1934). Contribution à l’étude de l’Apus cancriformis et de ses variations dans l’Afrique du Nord. Bull Soc Sci Nat Maroc 14: 125–139.
Ghigi A (1921). Ricerche sui Notostraci di Cirenaica a di altri paesi del Mediterraneo. Atti Soc Ital Sci Nat 60: 161–188.
Ghigi A (1924). Ancora sulla sistematica delle specie mediterranee del genere Triops. Atti Soc Ital Sci Nat 63: 193–202.
Gómez A, Carvalho GR (2000). Sex, parthenogenesis and genetic structure of rotifers: microsatellite analysis of contemporary and resting egg bank populations. Mol Ecol 9: 203–214.
Goudet J (2001). FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). (code available from http://www.unil.ch/izea/softwares/fstat.html).
Gurney R (1923). Notes on some British and North African specimens of Apus cancriformis, Schaeffer. Ann Mag Nat Hist 11: 496–502.
Kang TW, Lee EH, Kim MS, Paik SG, Kim S, Kim CB (2005). Molecular phylogeny and geography of Korean medaka fish (Oryzias latipes). Mol Cells 20: 151–156.
King JL, Hanner R (1998). Cryptic species in a ‘living fossil’ lineage: taxonomic and phylogenetic relationships within the genus Lepidurus (Crustacea: Notostraca) in North America. Mol Phylogenet Evol 10: 23–36.
Korn M, Marrone F, Pérez-Bote JL, Machado M, Cristo M, Cancela da Fonseca L et al. (2006). Sister species within the Triops cancriformis lineage (Crustacea, Notostraca). Zool Scr 35: 301–322.
Kumar S, Tamura K, Nei M (2004). MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163.
Linder F (1952). Contributions to the morphology and taxonomy of the Branchiopoda Notostraca, with special reference to the North American species. Proc US Nat Mus 102: 1–69.
Longhurst AR (1955). A review of the Notostraca. Bulletin of the British Museum (Natural History). Zoology 3: 1–57.
Mantovani B, Cesari M, Scanabissi F (2004). Molecular taxonomy and phylogeny of the ‘living fossil’ lineages Triops and Lepidurus (Branchiopoda: Notostraca). Zool Scr 33: 367–374.
Marescalchi O, Cesari M, Eder E, Scanabissi F, Mantovani B (2005). Chromosomes in sexual populations of notostracan and conchostracan taxa (Crustacea, Branchiopoda). Caryologia 58: 164–170.
Mathias P (1937). Biologie des Crustacés Phyllopodes. Hermann Editeurs: Paris.
Murugan G, Maeda-Martinez AM, Obrégon-Barboza H, Hernàndez-Saavedra NY (2002). Molecular characterization of the tadpole shrimp Triops (Branchiopoda: Notostraca) from the Baja California Peninsula, México: new insights on species diversity and phylogeny of the genus. Hydrobiologia 486: 101–113.
Pálsson S (2000). Microsatellite variation in Daphnia pulex from both sides of the Baltic Sea. Mol Ecol 9: 1075–1088.
Pannell JR (2002). The evolution and maintenance of androdioecy. Annu Rev Ecol Syst 33: 397–425.
Pfrender ME, Spitze K, Lehman N (2000). Multi-locus genetic evidence for rapid ecologically based speciation in Daphnia. Mol Ecol 9: 1717–1735.
Posada D, Crandall KA (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818.
Raymond M, Rousset F (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249.
Remigio EA, Hebert PDN (2000). Affinities among anostracans (Crustacea: Branchiopoda) families inferred from phylogenetic analyses of multiple gene sequences. Mol Phylogenet Evol 18: 117–128.
Sambrook E, Fritsch F, Maniatis T (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press: Cold Spring Harbor, NY.
Scanabissi F, Eder E, Cesari M (2005). Male occurrence in Austrian populations of Triops cancriformis (Branchiopoda, Notostraca) and ultrastructural observations of the male gonad. Invertebr Biol 124: 57–65.
Scanabissi F, Mondini C (2002). A survey of the reproductive biology in Italian branchiopods. Part B. The male gonad of Lepidurus apus lubbocki Brauer, 1873 (Notostraca). Hydrobiologia 486: 273–278.
Scott SR, Grigarick AA (1979). Laboratory studies of factors affecting egg hatch of Triops longicaudatus (LeConte) (Notostraca: Triopsidae). Hydrobiologia 63: 145–152.
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P (1994). Evolution weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87: 651–701.
Swofford DL (2001). PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0. Sinauer Associates: Sunderland, MA.
Templeton AR, Crandall KA, Sing CF (1992). A cladistic analysis of phenotipic association with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132: 619–633.
Trusheim F (1938). Triopsiden aus dem Keuper-Frankens. Palaeontol Z 19: 198–216.
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004). Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4: 535–538.
Vianna JA, Bonde RK, Caballero S, Giraldo JP, Lima RP, Clark A et al. (2006). Phylogeography, phylogeny and hybridization in trichechid sirenians: implications for manatee conservation. Mol Ecol 15: 433–447.
Weeks SC, Benvenuto C, Reed SK (2006). When males and hermaphrodites coexist: a review of androdioecy in animals. Integr Comp Biol 46: 449–464.
Wingstrand KG (1978). Comparative spermatology of the Crustacea Entomostraca. 1. Subclass Branchiopoda. Biol Skr Dan Vid Sel 22: 1–66.
Wright S (1969). Evolution and the genetics of populations. vol. 2: The Theory of Gene Frequencies. University of Chicago Press: Chicago.
Zaffagnini F, Trentini M (1980). The distribution and reproduction of Triops cancriformis (Bosc) in Europe (Crustacea, Notostraca). Monit Zool Ital 14: 1–8.
Acknowledgements
We are indebted to Dr Dani Boix, Dr Jordi Sala (Girona University) and Dr Loris Mularoni (IMIM, Barcelona) for collecting the Espolla specimens and to Dr Giuseppe Alfonso (Lecce University) for sampling the Lecce individuals. We also thank Dr Erich Eder (Wien University) for supplying the Austrian sample. This work was funded by MIUR 40% and Donazione Canziani (Bologna University) grants.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mantovani, B., Cesari, M., Luchetti, A. et al. Mitochondrial and nuclear DNA variability in the living fossil Triops cancriformis (Bosc, 1801) (Crustacea, Branchiopoda, Notostraca). Heredity 100, 496–505 (2008). https://doi.org/10.1038/hdy.2008.3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2008.3
Keywords
This article is cited by
-
Mitochondrial genome diversity and evolution in Branchiopoda (Crustacea)
Zoological Letters (2019)
-
Molecular and morphological delimitation of Australian Triops species (Crustacea: Branchiopoda: Notostraca)—large diversity and little morphological differentiation
Organisms Diversity & Evolution (2017)
-
R2 dynamics in Triops cancriformis (Bosc, 1801) (Crustacea, Branchiopoda, Notostraca): turnover rate and 28S concerted evolution
Heredity (2011)