Abstract
Purpose
The purpose of the present study is to develop fast automated quantification of retinal fluid in optical coherence tomography (OCT) image sets.
Methods
We developed an image analysis pipeline tailored towards OCT images that consists of five steps for binary retinal fluid segmentation. The method is based on feature extraction, pre-segmention, dimension reduction procedures, and supervised learning tools.
Results
Fluid identification using our pipeline was tested on two separate patient groups: one associated to neovascular age-related macular degeneration, the other showing diabetic macular edema. For training and evaluation purposes, retinal fluid was annotated manually in each cross-section by human expert graders of the Vienna Reading Center. Compared with the manual annotations, our pipeline yields good quantification, visually and in numbers.
Conclusions
By demonstrating good automated retinal fluid quantification, our pipeline appears useful to expert graders within their current grading processes. Owing to dimension reduction, the actual learning part is fast and requires only few training samples. Hence, it is well-suited for integration into actual manufacturer’s devices, further improving segmentation by its use in daily clinical life.
Similar content being viewed by others
Introduction
Spectral-domain optical coherence tomography (OCT) is nowadays the most frequently used retinal imaging technique in ophthalmology. It provides good visualization of subretinal and intraretinal cystoid fluid. Especially distinguishing between fluid and no fluid or a fluid increase or decrease between two visits is important for the administration of anti-vascular endothelial growth factor therapy. However, automated quantification of retinal fluid in OCT images goes beyond evaluation algorithms provided by manufacturers. When considering that a regular OCT raster scan has between 128 and 1024 B-scans, this can be tedious and beyond possibilities of every day clinical routine (cf. Schmidt-Erfuth et al1).
Owing to increased computing power, state-of-the-art methods for several learning tasks related to image analysis in the applied sciences are deep neural networks (NNs).2 Deep convolutional NNs, in particular, have already been applied to fluid quantification in OCT images (cf. Schlegl3). Deep learning models have several drawbacks, such as the need for huge amounts of training data, high training time, large memory usage, and the complicated design of a suitable network. A semi-supervised technique was developed by Zheng et al.4 (see also Chen et al,5 Xu et al,6 and Wilkins et al;7 for general learning techniques, we refer to Grady,8 Czaja and Ehler,9 Schölkopf and Smola,10 and Roweis and Saul11). Here we shall provide an efficient method with a fast learning part and low memory needs based on dimension reduction techniques. Moreover, it shall be easy to implement, requires only few training data, and yields good fluid quantification. To meet clinical needs, we present a fully automated image analysis pipeline tailored to fluid quantification in OCT raster image sets. The pipeline is built by five steps that are guided by the principle of dimension reduction. Our resulting binary segmentations are compared with the manual annotations of the expert graders visually and in numbers, illustrating the strengths and weaknesses of our approach. The innovative aspect of our approach is the combination of local and global features in dimension reduction techniques together with learning methods. Our approach presents a model applicable for clinical routine and could be implemented into a OCT device for its every day use.
Methods
The method is designed for a collection of 3D OCT volumes. For training and evaluation purposes, retinal fluid was annotated manually in each cross-section by human expert graders of the Vienna Reading Center (VRC). Guaranteeing unbiased evaluation, the data were annotated before the development of the pipeline and hence independently analyzed. As manual annotation is very time consuming (~15 h per volume), there are few training data available, that is, binary annotations of retinal fluid in few cross-sections, which enable the application of supervised learning techniques.
Our image analysis pipeline is built up from five steps. To reduce computation costs and minimize required training data, we focus on the extraction of the necessary image information contents before entering the learning stages. More specifically, (1) we compute local features for each B-scan that enhance specific image characteristics, while inheriting effective denoising. For each OCT B-scan, we compute two local features. One results from Gabor filtering12 and the other one as the solution of a variational minimization problem13 resulting in a piecewise smooth image. These two local features form a vector-valued image (cf. Local features). (2) We replace each B-scan with the piecewise constant solution of another variational energy minimization problem,14 whose input is the vector-valued image. This solution is considered as a pre-segmentation, in which darker piecewise constant areas appear to be associated with retinal fluid (cf. Dimension reduction I: pre-segmentation). We now aim to select those areas by automated learning an appropriate threshold for each cross-section, such that segments with pixel values underneath the threshold represent the fluid. To minimize computation costs and reduce the required training data in the learning process, (3) we compute four global parameters of each cross-section (cf. Dimension reduction II). (4) We now apply three standard learning tools (regression, decision trees, and NNs) to estimate the thresholds from these parameters based on few randomly selected manual annotations of expert graders serving as training samples (cf. Supervised learning of the thresholds). We then evaluate the trained learning models on the global parameters of each B-scan of the remaining test samples. (5) Once the thresholds are estimated, we use them to threshold the piecewise constant pre-segmentation images obtaining the final identification of retinal fluid areas (cf. Thresholding). Steps 2 and 3 are significant dimension reduction processes enabling faster learning with fewer training data. We run through these five steps for each 2D cross-sectional image independently, yielding the following analysis pipeline:
-
Local features (pixel-level):Compute two features for each B-scan, yielding vector-valued images.
-
Dimension reduction I (pre-segmentation):Use these vector-valued images to replace each B-scan with the piecewise constant solution of an energy minimization problem.
-
Dimension reduction II:Represent each B-scan by few global features (image level).
-
Learning thresholds:From these global features estimate the thresholds via standard learning tools.
-
Thresholding:Each piecewise constant B-scan is thresholded to obtain the final classification of the fluid spaces.
Local features
We extract two local features of each B-scan (see the first two images in Figure 1). One feature image is obtained from Gabor filtering (see Supplementary Equation 1). Gabor filters are well-suited for texture analysis;12 see also Soares et al15 for applications in retinal image analysis. The second feature is obtained from the Rudin–Osher–Fatemi variational energy minimization13 (see Supplementary Equations 2 and 3). Its solution is a piecewise smooth image, that is, edges are preserved and regions between interfaces are smoothed.
Dimension reduction I: pre-segmentation
We now use the vector-valued feature images to replace each B-scan with the piecewise constant solution of another variational energy minimization problem, (see Supplementary Equation 4 and cf. Mumford and Shah,16 and Potts.17 We minimize a discrete domain version (see Supplementary Equation 5) by using the Pottslab matlab code from the website http://www.pottslab.de (cf. Storath and Weinmann14), where an alternating direction method of multipliers is used.
It is noteworthy that the minimizer is again a vector-valued image. We use the second channel of the piecewise constant image, which corresponds to the Rudin–Osher–Fatemi model, see Figure 2. Indeed it yields pre-segmentations in which retinal fluid appears as dark regions and boundaries seem preserved well. Thus, it seems reasonable to threshold the piecewise constant image to divide the piecewise constant images in two regions: white (presence of retinal fluid) and black (absence). As annotations from human graders are available, supervised learning techniques can be used to estimate the magnitude of appropriate thresholds.
Dimension reduction II
In order to learn the thresholds, we need to specify the parameters of the piecewise constant image that actually characterize the thresholds magnitude. We observed that the threshold is mainly influenced by few global image parameters (mean, variance, infimum, and median). Therefore, we associate just these four global features of the region of interest (ROI) with each piecewise constant image that are used as input for subsequent learning.
Supervised learning of the thresholds
We split our cross-sections into training and test samples. As training data, we use few randomly chosen 2D slices and test it on all remaining cross-sections. First, we manually determine thresholds ti∈[0, 1] as target values, based on the optimal fit with the graders’ annotations.
To estimate thresholds from four global features (see Dimension reduction II), we make use of three learning techniques: regression, random forest, and three-layer NNs (cf. Bishop,18 and Witten and Frank19 for more details). The target thresholds {ti} are used to train these learning models. We obtain the remaining thresholds for the test data set, which is disjoint from the training data, by evaluating the trained learning model on the global features of the test samples. The machine learning and NN toolbox of MatlabR2015a is used for the learning computations.
Thresholding
The last step of our pipeline is to threshold all the computed pre-segmentations (see Dimension reduction I: pre-segmentation) of the test set by the evaluated thresholds. Those segments with values less than the threshold are classified as retinal fluid (see right image in Figure 1).
Test data
Our proposed analysis pipeline is tested on imaging data provided by the VRC. The VRC has analyzed millions of images, mainly from OCT, recorded in multi-center clinical trials with many hundred different study sites all over the world in a standardized manner over the past decade. We shall work here with two separate OCT data sets, the first corresponds to 38 patients suffering from neovascular age-related macular degeneration (AMD), where OCT images are taken at the very start of the treatment initiation (ie, all are baseline images). The second data set relates to 16 patients suffering from diabetic macular edema (DME) with different severity of edema selected randomly from a trial database within the VRC. All OCT images were recorded by a Cirrus OCT camera (Carl Zeiss Meditec, Dublin, CA, USA). We will test the pipeline on both data sets independently.
Results
For each patient of the two data sets, the recorded retinal image data yields a three-dimensional (1024 × 512 × 128) matrix, that is, the volume data consists of 128 cross-sectional images of one patient’s retina of one eye each of the size (1024 × 512). We cut out scaled images of the size (512 × 512) that include the ROI, that is, the region between the inner limiting membrane and the pigment epithelium cells, which is the retina (see Garvin et al20 and Haeker et al21 for retinal layer segmentation). This halves the amount of pixels of each 2D image without loosing any information. Retinal fluid was annotated in each of the 4864=38*128 and 2048=16*128 cross-sections manually by human expert graders of the VRC, provided as binary image sets. Using professional drawing tablets, all annotations were performed B-scan-wise. The time required to annotate a typical OCT volume was ~15 h. In total, five readers and three board-certified supervisors, who corrected errors if required, participated in the annotation procedure. Each reader underwent a standardized training before annotating the study data set (see Waldstein et al22 and Varnousfaderani et al23 for more details).
The results of our fully automated quantification of subretinal and intraretinal cystoid fluid highly depend on the quality of the piece-wise constant pre-segmentation. Table 1 shows good binary segmentation outcomes for both data sets when we use the optimal threshold on the piece-wise constant training data and compare it with the graders’ annotations. It is noteworthy that sensitivity, specificity, and dice coefficients from the training sets (see Table 1) serve as the baselines for evaluations on the test data.
Qualitative/visual evaluation
In Figure 3, we show original OCT images on the left, next to the same images with the retinal fluid annotated by human expert graders in the center. On the right, we provide the output of our image analysis pipeline, which is our resulting binary segmentation. In these selected cases, retinal fluid locations are relatively unambiguous and graders’ annotations are consistent with the visual impressions. Our segmentations coincide well with the graders’ annotations.
The human expert graders annotate each cross-section separately, which leads to inconsistencies in few subsequent cross-sections that appear almost identical. In Figure 4a, we see that similarly looking cross-sections are annotated differently by human graders, whereas our algorithm identifies retinal fluid consistently in both cases. Which is clinically correct in this case remains unclear, as the fluid can be hidden by other pathologic structures and the human annotations always leave a little room for subjectiveness and interpretation, even when high standards for objectivity are set. In any case, a consistent result is important and this can only be achieved by the algorithm. Retinal vessels cast shadows in OCT, attenuating the signal underneath and yielding darker regions in the original images. It should be noticed that, without further substeps involved, such regions get marked incorrectly by our pipeline (cf. Figure 4b).
Some challenges of the processing: (a) similar looking images are annotated differently by human graders, whereas our algorithm identifies retinal fluid consistently in both cases. Row 1 and 2: (left) original image, (center) graders’ annotations, and (right) pipeline output. (b) Shadows from vessels are marked wrong by our pipeline. Row 3: (left) original image, (center) graders’ annotations, and (right) pre-segmentation.
Quantitative evaluation
In Table 1, we compare our resulting binary segmentations with the manual annotations of the expert graders serving as the reference of quality.
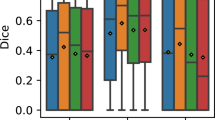
Two types of quantitative evaluations are considered. We refer to classification when we aim to detect cross-sections that contain retinal fluid, that is, each 2D cross-section is globally classified into either presence or absence of retinal fluid. We refer to volume quantification when comparing the pipelines output in the ROI pixel-wise with the graders’ annotations in each cross-section. Classification and volume quantification are compared for the two data sets separately. Comparing the three different learning techniques sensitivity and specificity for classification are at the same level. The NNs are above the 70% bound for both data sets. For volume quantification, regression lacks behind on the DME data. NNs, on the other hand, yield good success rates with dice coefficients bigger than 0.5 for both data sets.
We also provide mean and variance difference between the manual annotations and our quantified retinal fluid volume in mm3, where camera specifications yield that one pixel translates to ≈10−6 mm3. The corresponding Bland–Altmann plot in Figure 1 of the Supplementary Material shows reasonable agreement between annotations and computed segmentation for the volume quantification using the NNs.
Computational resources
The run-time in MatlabR2015a for one OCT volume (128 B-Scans) on a 2.2 GHz Intel Core i7 processor with 16 GB RAM is ~45 min. The vector-valued minimization (Supplementary Equation 5) is the most time-consuming part (43 min), which is independent from the actual learning taking less than a second.
Discussion
The development of our pipeline was guided by the idea of dimension reduction in order to simplify the actual learning part. Although the pre-segmentation is the first dimension reduction by significantly quantizing the intensity range of the image, the most extensive dimension reduction stems from learning the thresholds from four global features per cross-section. It seems that the threshold effectively only depends on global statistics of the pre-segmentation, enabling cheap and fast learning. The second step covers the essential image information. Gabor filtering takes care of texture and the Rudin–Osher–Fatemi model enhances edges, while denoising between interfaces. The subsequent piecewise constant energy minimization yields a pre-segmentation and is the basis for final binary segmentation through thresholding. We have seen that this pre-segmentation is sufficiently rich, so that optimal thresholding on the training data yields regions that coincide well with the provided manual annotations. The pre-segmentation is time-consuming relative to the other pipeline steps. Faster pre-segmentation methods are availabe but the piecewise constant minimizer of (Supplementary Equation 4) yields the best results for our concerns at the moment.
There is a beneficial interaction between features, pre-segmentation, dimension reduction, and threshold learning, which yields convincing results. For each patient set, we used only 100 randomly selected B-scans as training samples for evaluating the pipeline on the remaining cross-sections yielding 4764 (AMD) and 1948 (DME) test samples. It is a big benefit that only few manually annotated training data are needed in our pipeline, as large amounts of manual annotations are often not available. Our image analysis pipeline for quantification of retinal fluid in OCT has the potential to significantly support human expert graders. By our automated classification into presence or absence of retinal fluid, graders can disregard a large subset of B-scans speeding up the manual grading process. Our results for the detection yield sufficient sensitivity and specificity rates, so that its integration into the grading process seems useful. Especially considering the growing number of B-scans and details in each B-scan due to higher resolution in all scan directions, automated analysis methods will be needed in ophthalmology. Here, it is of greatest importance to have semi-automated segmentation tools for the validation of proposed automated methods. Our method will speed up the annotation in a reading center setting by giving a fast first attempt as the basis for manual annotation correction, or within the frame of clinical routine, giving the ophthalmologist a quick first result for the interpretation of the scan. When compared with other commonly used computerized evaluation in biomedicine, such as automated interpretation of electrocardiograms, our pipeline appears definitely competitive (cf. NAMEstes24 and Wetherell25).
Concerning our automated quantification of retinal fluid, we saw in Results that visual comparison of our results with the graders’ annotations appear convincing. For high-quality manual annotations, our segmented regions coincide well. However, earlier studies have shown that inconsistencies between graders are normal to a certain percentage and we can only aim at the same inter-grader reproducibility such as the reported human inter-grader variability (cf. Varnousfaderani23). In Figure 4a, cross-sections appear similar but graders’ annotations differ. Medical experts confirm inconsistency and our automated approach delivers consistent segmentations in that case. Taking away subjectivity by using automated segmentation is of particular importance when it comes to tracking retinal fluid changes in patients over time. Such deviations from the annotations also contribute to the error rates in the quantitative evaluation (Table 1). Further falsely segmented areas are due to vessel shadows, which can easily be removed by human graders’ when our pipeline is used as a supporting tool as indicated above. For a fully automated quantification, vessel shadows should be taken care of in additional processing steps (cf. Girard et al,26 Wu et al,27 and Ehler et al).28 When our pipeline is tested on a larger consistently annotated data set and adapted to the removal of false positives due to vessel shadowing, it could be used as an analysis tool in an OCT device. Many of today’s device algorithms (eg, for retinal thickness segmentation) can show errors in severe pathologic cases and clinicians’ feedback corrections of the device software are needed. Owing to our fast learning time, we can even envision to improve our algorithm by using segmentation corrections of clinicians for new learning sets centrally at the device manufacturer further improving by its use in daily clinical life.
In comparison with other automated retinal fluid quantification attempts in the literature, our pipeline’s learning step is exceptionally efficient due to dimension reduction. In Chen et al,5 k-nearest neighbor learning with ≈50 dimensional feature vectors is combined with graph cuts for voxel-based fluid segmentation. Our dimension reduction methodology combined with learning is pixel based and requires only two features. In contrast to Xu et al,6 we do not require a full retinal layer segmentation and reasonably smooth fluid boundaries are guaranteed by thresholding our specific pre-segmentation. As opposed to Zheng et al,4 we do not require any human interaction during the analysis procedure. Cystoid macular edema has been identified in Wilkins et al7 by an automated segmentation based on bilateral filtering and thresholding. The bilateral filtering shares similarities to the Rudin–Osher–Fatemi model, but we do not directly threshold; instead, we make a detour by computing a reasonable pre-segmentation. The algorithm in Wilkins et al7 uses orders of magnitude more training data and is more empirically tailored to their specific data set. Their test data are few manually selected cross-sections and the results were evaluated afterwards manually, so that there seems a chance that the expert annotations are not fully unbiased from the computational results. Our pipeline works well on two independent data sets with different retinal diseases and we just use about 2 and 5% of the data sets for training, respectively. Any potential bias in graders’ annotations from our computational results is avoided in the first place as the pipeline development was started after the annotations were performed. In addition, the VRC, as an independent and unbiased institution for the standardized evaluation of many multicenter clinical trials (including drug licensing trials) is used to blinding of image graders and supervisors to the purpose of their evaluation when needed.
Deep convolutional NNs have been successfully applied to detect subretinal and intraretinal cystoidal fluid.3 However, deep learning requires large amounts of training data. In Lee et al,29 central OCT scans are linked to electronic medical records enabling the application of deep convolutional NNs. There, 80 839 training samples are used to distinguish between normal and AMD scans in 20 163 validation samples. Beyond classification, our target is fluid quantification. As the amount of accurate pixel-wise fluid annotations is often limited, we aimed to learn efficiently and use only 100 training and 4764 validation samples in AMD patients.
In conclusion, we have demonstrated that our image analysis pipeline yields good automated retinal fluid quantification in OCT images and our visual results could be useful to expert graders within their current grading processes. The pipeline is based on dimension reduction techniques with a fast learning part and low memory needs. Furthermore, it requires exceptionally few training data and is sufficiently versatile for further adjustments in the sense that current steps allow for modifications and additional steps can easily be inserted. The successful interplay of enormous dimension reduction with relatively elementary learning techniques is remarkable.
We envision several aspects for future work; for instance, we will incorporate artifact removal from vessel shadows by integrating ideas from Girard et al26, 27 and Wu et al27 Moreover, so far, graders at the VRC annotated each OCT cross-section separately, dismissing the full spatial correlations of the 3D volumes. All of our pipeline steps are either already available or can be implemented in higher dimensions, so that we plan to perform segmentation in 3D directly. However, suitable 3D annotations are needed (cf. Figure 4a), but are not easy to consistently annotate even for an experienced grader or retinal specialist. Both, vessel shadow removal and 3D implementations are work in progress.

References
Schmidt-Erfurth U, Klimscha S, Waldstein SM, Bogunovic H . A view of the current and future role of optical coherence tomography in the management of age-related macular degeneration. Eye 2017; 31: 26–44.
LeCun Y, Bengio Y, Hinton G . Deep learning. Nature 2015; 521: 436–444.
Schlegl T, Glodan A, Podkowinski D, Waldstein SM, Gerendas BS, Schmidt-Erfurth U et al. Automatic segmentation and classification of intraretinal cystoid fluid and subretinal fluid in 3D-OCT using convolutional neural networks. Invest Ophthalmol Vis Sci 2015; 560 (7).
Zheng Y, Sahni J, Campa C, Stangos AN, Raj A, Harding SP . Computerized assessment of intraretinal and subretinal fluid regions in spectral-domain optical coherence tomography images of the retina. Am J Ophthalmol 2013; 155: 277–286.
Chen X, Niemeijer M, Zhang L, Lee K, Abramoff MD, Sonka M . 3D segmentation of fluid-associated abnormalities in retinal OCT: probability constrained graph-search—graph-cut. IEEE Trans Med Imaging 2012; 31 (8): 1521–1531.
Xu X, Lee K, Zhang L, Sonka M, Abramoff MD . Stratified sampling voxel classification for segmentation of intraretinal and subretinal fluid in longitudinal clinical OCT data. IEEE Trans Med Imaging 2015; 340 (7): 1616–1623.
Wilkins G, Houghton O, Oldenburg A . Automated segmentation of intraretinal cystoid fluid in optical coherence tomography. IEEE Trans Biomed Eng 2012; 59 (4): 1109–1114.
Grady L . Random walks for image segmentation. IEEE Trans Pattern Anal Mach Intell 2006; 28 (11): 1768–1783.
Czaja W, Ehler M . Schrödinger Eigenmaps for the analysis of biomedical data. IEEE Trans Pattern Anal Mach Intell 2013; 35 (5): 1274–1280.
Schölkopf B, Smola A . Learning with Kernels. MIT Press: Cambridge, MA, 2002.
Roweis ST, Saul LK . Nonlinear dimensionality reduction by locally linear embedding. Science 2000; 290 (5500): 2323–2326.
Daugman J . Complete discrete 2-D Gabor transforms by neural networks for image analysis and compression. IEEE Trans Acoust 1988; 360 (7): 1169–1179.
Rudin L, Osher S, Fatemi E . Nonlinear total variation based noise removal algorithm. Phys D 1992; 60: 259–268.
Storath M, Weinmann A . Fast partitioning of vector-valued images. SIAM J Imaging Sci 2014; 7: 1826–1852.
Soares JVB, Leandro JJG, Cesar RM, Jelinek HF, Cree MJ . Retinal vessel segmentation using the 2-D Gabor wavelet and supervised classification. IEEE Trans on Med Imaging 2006; 25 (9): 1214–1222.
Mumford D, Shah J . Optimal approximations by piecewise smooth functions and associated variational problems. Comm Pure Appl Math 1989; 420 (5): 577–685.
Potts R . Some generalized order-disorder transformations. Proc Cambridge Philos Soc 1952; 48: 106–109.
Bishop CM . Pattern Recognition and Machine Learning. Springer-Verlag: New York, 2006.
Witten IH, Frank. E . Data Mining: Practical Machine Learning Tools and Techniques. Elsevier, 2005.
Garvin MK, Abramoff MD, Wu X, Russell SR, Burns TL, Sonka M . Automated 3-D intraretinal layer segmentation of macular spectral-domain optical coherence tomography images. IEEE Trans Med Imaging 2009; 28 (9): 1436–1447.
Haeker M, Abramoff M, Kardon R, Sonka M . Segmentation of the surfaces of the retinal layer from OCT images. Med Image Comput Comput Assist Interv 2006; 9 (Pt 1): 800–807.
Waldstein SM, Philip A-M, Leitner R, Simader C, Langs G, Gerendas BS et al. Correlation of 3-dimensionally quantified intraretinal and subretinal fluid with visual acuity in neovascular age-related macular degeneration. JAMA Ophthalmol 2016; 134 (2): 182–190.
Varnousfaderani ES, Wu J, Vogl W-D, Philip A-M, Montuoro A, Leitner R et al. A novel benchmark model for intelligent annotation of spectral-domain optical coherence tomography scans using the example of cyst annotation. Comput Methods Programs Biomed 2016; 130: 93–105.
Estes NAM . Computerized interpretation of ECGs. Circ Arrhythm Electrophysiol 2013; 6: 2–4.
Wetherell H . Can we trust our ECG machines? Br J Cardiol 2014; 21: 62–63.
Girard MJA, Strouthidis NG, Ethier CR, Mari JM . Shadow removal and contrast enhancement in optical coherence tomography images of the human optic nerve head. Invest Ophthalmol Vis Sci 2011; 52 (10): 7738–7748.
Wu J, Gerendas BS, Waldstein SM, Simader C, Schmidt-Erfurth U . Automated vessel shadow segmentation of fovea-centered spectral-domain images from multiple OCT devices. Proc. SPIE 9034, Medical Imaging 2014; 9034: 903403.
Ehler M, Dobrosotskaya J, Cunningham D, Wong WT, Chew EY, Czaja W et al. Modeling photo-bleaching kinetics to create high resolution maps of rod rhodopsin in the human retina. PLoS One 2015; 10 (7): e0131881.
Lee CS, Baughman DM, Lee. AY . Deep learning is effective for classifying normal versus age-related macular degeneration optical coherence tomography images. Ophthalmol Retina, in press 2017.
Acknowledgements
This work was funded by the Vienna Science and Technology Fund (WWTF) through project VRG12-009.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Eye website
Supplementary information
Rights and permissions
About this article
Cite this article
Breger, A., Ehler, M., Bogunovic, H. et al. Supervised learning and dimension reduction techniques for quantification of retinal fluid in optical coherence tomography images. Eye 31, 1212–1220 (2017). https://doi.org/10.1038/eye.2017.61
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/eye.2017.61
This article is cited by
-
Deep learning approach based on dimensionality reduction for designing electromagnetic nanostructures
npj Computational Materials (2020)
-
On Orthogonal Projections for Dimension Reduction and Applications in Augmented Target Loss Functions for Learning Problems
Journal of Mathematical Imaging and Vision (2020)
-
Künstliche Intelligenz zum Management von Makulaödemen
Der Ophthalmologe (2020)