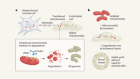
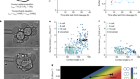
- NEWS AND VIEWS
One ring to rule them all
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 575, 291-293 (2019)
doi: https://doi.org/10.1038/d41586-019-03200-4
References
Zhang, Y. et al. Nature 573, 600–604 (2019).
Zhang, X. et al. Nature 575, 385–389 (2019).
Rao, S. S. et al. Cell 159, 1665–1680 (2014).
Ji, Y. et al. Cell 141, 419–431 (2010).
Hu, J. et al. Cell 163, 947–959 (2015).
Jain, S., Ba, Z., Zhang, Y., Dai, H. Q. & Alt, F. W. Cell 174, 102–116 (2018).
Dong, J. et al. Nature 525, 134–139 (2015).
Seitan, V. C. et al. Nature 476, 467–471 (2011).
Rao, S. S. P. et al. Cell 171, 305–320 (2017).
Canela, A. et al. Cell 170, 507–521 (2017).

 Read the paper: Fundamental roles of chromatin loop extrusion in antibody class switching
Read the paper: Fundamental roles of chromatin loop extrusion in antibody class switching
 Read the paper: The fundamental role of chromatin loop extrusion in physiological V(D)J recombination
Read the paper: The fundamental role of chromatin loop extrusion in physiological V(D)J recombination
 Pressure regulates immune-cell function
Pressure regulates immune-cell function
 Unequal opportunity during class switching
Unequal opportunity during class switching