- NEWS AND VIEWS
Tiny crystals have big potential for determining structures of small molecules
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 564, 348-349 (2018)
doi: https://doi.org/10.1038/d41586-018-07756-5
References
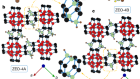
Gruene, T. et al. Angew. Chemie Int. Edn 57, 16313–16317 (2018).
Jones, C. G. et al. ACS Central Sci. 4, 1587–1592 (2018).
Shi, D., Nannenga, B. L., Iadanza, M. G. & Gonen, T. eLife 2, e01345 (2013).
Mugnaioli, E., Gorelik, T. & Kolb, U. Ultramicroscopy 109, 758–765 (2009).
Su, J. et al. Microporous Mesoporous Matter 189, 115–125 (2014).
Zerikly, M. & Challis, G. L. ChemBioChem 10, 625–633 (2009).
Gross, L., Mohn, F., Moll, N., Liljeroth, P. & Meyer, G. Science 325, 1110–1114 (2009).
Inokuma, Y. et al. Nature 495, 461–466 (2013).

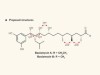 Molecular structure assignment simplified
Molecular structure assignment simplified
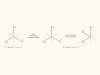 Handedness detected by microwaves
Handedness detected by microwaves