- NEWS AND VIEWS
All for one and one for all to fight flu
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 565, 29-31 (2019)
doi: https://doi.org/10.1038/d41586-018-07654-w
References
Morens, D. M. & Fauci, A. S. J. Infect. Dis. 195, 1018–1028 (2007).
Paules, C. & Subbarao, K. Lancet 390, 697–708 (2017).
Laursen, N. S. et al. Science 362, 598–602 (2018).
Taubenberger, J. K. & Kash, J. C. Cell Host Microbe 7, 440–451 (2010).
Ekiert, D. C. et al. Science 324, 246–251 (2009).
Sui, J. et al. Nature Struct. Mol. Biol. 16, 265–273 (2009).
Corti, D. et al. Science 333, 850–856 (2011).
Margine, I. & Krammer, F. Pathogens 3, 845–874 (2014).
Jegaskanda, S., Reading, P. C. & Kent, S. J. J. Immunol. 193, 469–475 (2014).
Nachbagauer, R. & Krammer, F. Clin. Microbiol. Infect. 23, 222–228 (2017).
Peyvandi, F. et al. N. Engl. J. Med. 374, 511–522 (2016).
Borsotti, C. & Follenzi, A. Expert Rev. Clin. Immunol. 14, 1013–1019 (2018).
Hai, R. et al. J. Virol. 86, 5774–5781 (2012).
Impagliazzo, A. et al. Science 349, 1301–1306 (2015).
Yassine, H. M. et al. Nature Med. 21, 1065–1070 (2015).
Xu, L. et al. Science 358, 85–90 (2017).
Competing Interests
G.J.N. and J.W.S. are employees of Sanofi, whose Sanofi-Pasteur vaccine subsidiary develops and manufactures influenza vaccines and whose Ablynx subsidiary develops nanobody medicines. G.J.N. is chief scientific officer and J.W.S. is director of Sanofi-Pasteur vaccine research and development.
G.J.N. is listed on intellectual property developed and owned by the National Institutes of Health that relates to the development of novel influenza vaccines.

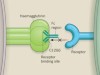 A broadly protective antibody
A broadly protective antibody
 Prediction is worth a shot
Prediction is worth a shot