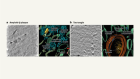
- TECHNOLOGY FEATURE
Cryo-electron microscopy shapes up
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 561, 565-567 (2018)
doi: https://doi.org/10.1038/d41586-018-06791-6
References
Renaud, J. P. et al. Nature Rev. Drug Discov. 17, 471–492 (2018).
Subramaniam, S., Earl, L. A., Falconieri, V., Milne, J. L. S. & Egelman, E. H. Curr. Opin. Struct. Biol. 41, 194–202 (2016).
Neumann, P., Dickmanns, A. & Ficner, R. Structure 26, 785–795.e4 (2018).
Wlodawer, A., Li, M. & Dauter, Z. Structure 25, 1589–1597.e1 (2017).
Heymann, J. B. et al. J. Struct. Biol. https://doi.org/10.1016/j.jsb.2018.08.010 (2018).
Afonine, P. V. et al. Acta Cryst. D 74, 814–840 (2018).
Herzik, M. A. Jr, Fraser, J. & Lander, G. C. Preprint at bioRxiv https://doi.org/10.1101/128561 (2017).
Joseph, A. P., Lagerstedt, I., Patwardhan, A., Topf, M. & Winn, M. J. Struct. Biol. 199, 12–26 (2017).

 The revolution will not be crystallized: a new method sweeps through structural biology
The revolution will not be crystallized: a new method sweeps through structural biology
 Cryo-electron microscopy wins chemistry Nobel
Cryo-electron microscopy wins chemistry Nobel
 The field that came in from the cold
The field that came in from the cold