Abstract
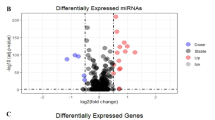
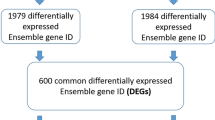
To identify the differentially expressed genes (DEGs) and their transcription factors (TFs) in colorectal cancer (CRC). We performed an integrated analysis of microarray studies. Functional annotation and CRC-specific transcriptional regulatory network construction were performed. Expression of selected DEGs and TFs was verified with The Cancer Genome Atlas (TCGA) data sets and qRT-PCR. SurvMicro was used to analyze the correlation between the overall survival time of CRC patients and the expression of DEGs and TFs. Seven data sets were obtained and 2014 DEGs in CRC were identified. Pathways in cancer and fatty acid metabolism were significantly enriched pathways of upregulated and downregulated DEGs, respectively. Expression of five DEGs (RERGL, ESM1, CA1, ANGPTL7 and TMEFF2) and their five TFs (ZNF354C, ARID3A, NFIC, BRCA1 and ZEB1) was verified by TCGA data sets and qRT-PCR. Their expression in TCGA data sets was same as that in our integrated analysis. Only the expression of EMS1, NFIC, BRCA1 and ZEB1 was inconsistent with integrated analysis and TCGA data sets. Expression of RERGL and BRCA1 was significantly correlated with the overall survival time of CRC patients. These five DEGs may have roles in CRC regulated by their five upstream TFs, which may make a contribution in uncovering the mechanism and providing new strategy of diagnosis and therapies for CRC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM . Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–2917.
Yang R, Chen B, Pfütze K, Buch S, Steinke V, Holinski-Feder E et al. Genome-wide analysis associates familial colorectal cancer with increases in copy number variations and a rare structural variation at 12p12.3. Carcinogenesis 2014; 35: 315–323.
Nakayama T, Hirakawa H, Shibata K, Nazneen A, Abe K, Nagayasu T et al. Expression of angiopoietin-like 4 (ANGPTL4) in human colorectal cancer: ANGPTL4 promotes venous invasion and distant metastasis. Oncol Rep 2011; 25: 929–935.
Su CY, Lin TC, Lin YF, Chen MH, Lee CH, Wang HY et al. DDX3 as a strongest prognosis marker and its downregulation promotes metastasis in colorectalcancer. Oncotarget 2015; 6: 18602–18612.
Rudolph A, Shi H, Forsti A, Hoffmeister M, Sainz J, Jansen L et al. Repeat polymorphisms in ESR2 and AR and colorectal cancer risk and prognosis: results from a German population-based case–control study. BMC Cancer 2014; 14: 817.
Wang ZS, Cong ZJ, Luo Y, Mu YF, Qin SL, Zhong M et al. Decreased expression of interleukin-36alpha predicts poor prognosis in colorectal cancer patients. Int J Clin Exp Pathol 2014; 7: 8077–8081.
Li Y, Li Y, Chen W, He F, Tan Z, Zheng J et al. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget 2015; 6: 27641–27650.
Kunizaki M, Sawai T, Takeshita H, Tominaga T, Hidaka S, To K et al. Clinical value of serum p53 antibody in the diagnosis and prognosis of colorectal cancer. Anticancer Res 2016; 36: 4171–4175.
Letellier E, Schmitz M, Baig K, Beaume N, Schwartz C, Frasquilho S et al. Identification of SOCS2 and SOCS6 as biomarkers in human colorectal cancer. Br J Cancer 2014; 111: 726–735.
Lee KW, Lee SS, Kim SB, Sohn BH, Lee HS, Jang HJ et al. Significant association of oncogene YAP1 with poor prognosis and cetuximab resistance in colorectal cancer patients. Clin Cancer Res 2015; 21: 357–364.
Ahmadi S, Sharifi M . Locked nucleic acid inhibits miR-92a-3p in human colorectal cancer, induces apoptosis and inhibits cell proliferation. Cancer Gene Ther 2016; 23: 199–205.
Akao Y, Nakagawa Y, Hirata I, Iio A, Itoh T, Kojima K et al. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther 2010; 17: 398–408.
Zhang X, Zhang Y, Yang J, Li S, Chen J . Upregulation of miR-582-5p inhibits cell proliferation, cell cycle progression and invasion by targeting Rab27a in human colorectal carcinoma. Cancer Gene Ther 2015; 22: 475–480.
Wang R, Du L, Yang X, Jiang X, Duan W, Yan S et al. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J Cancer Res Clin Oncol 2016; 142: 2291–2301.
He Q, Chen HY, Bai EQ, Luo YX, Fu RJ, He YS et al. Development of a multiplex MethyLight assay for the detection ofmultigene methylation in human colorectal cancer. Cancer Genet Cytogenet 2010; 202: 1–10.
Kondo Y, Issa JPJ . Epigenetic changes in colorectal cancer. Cancer Metastasis Rev 2004; 23: 29–39.
Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014; 63: 317–325.
Pedersen SK, Baker RT, McEvoy A, Murray DH, Thomas M, Molloy PL et al. A two-gene blood test for methylated DNA sensitive for colorectal cancer. PLoS ONE 2015; 10: e0125041.
Lanza G, Ferracin M, Gafà R, Veronese A, Spizzo R, Pichiorri F et al. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer 2007; 6: 1–11.
Zhao H, Cai W, Su S, Zhi D, Lu J, Liu S . Screening genes crucial for pediatric pilocytic astrocytoma using weighted gene coexpression network analysis combined with methylation data analysis. Cancer Gene Ther 2014; 21: 448–455.
Xiao WH, Qu XL, Li XM, Sun YL, Zhao HX, Wang S et al. Identification of commonly dysregulated genes in colorectal cancer by integrating analysis of RNA-Seq data and qRT-PCR validation. Cancer Gene Ther 2015; 22: 278–284.
Xiong Y, Wu S, Du Q, Wang A, Wang Z . Integrated analysis of gene expression and genomic aberration data in osteosarcoma (OS). Cancer Gene Ther 2015; 22: 524–529.
Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M et al. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res 2002; 62: 233–240.
Clarke PA, Poele RT, Wooster R, Workman P . Gene expression microarray analysis in cancer biology, pharmacology, and drug development: progress and potential 2. Biochem Pharmacol 2001; 62: 1311–1336.
Koga Y, Yamazaki N, Takizawa S, Kawauchi J, Nomura O, Yamamoto S et al. Gene expression analysis using a highly sensitive DNA microarray for colorectal cancer screening. Anticancer Res 2014; 34: 169–176.
Bianchini M, Levy EM, Zucchini C, Pinsky V, Macagno C, Valvassori L et al. Analysis of gene expression in human colorectal cancer tissues by cDNA microarray: implications for novel immunotherapy strategies. Cancer Res 2005; 65: 841-.
Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J et al. Metastasis-associated gene expression changes predict poor outcomes in patients with dukes stage B and C colorectal cancer. Clin Cancer Res 2009; 15: 7642–7651.
Andersen CL, Thorsen K, Kruhøffer M, Ørntoft TF . Transcription factors dysregulated in colorectal cancer. Cancer Res 2006; 66: 407-.
Kryczek I, Lin Y, Nagarsheth N, Peng D, Zhao L, Zhao E et al. IL-22+CD4+T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity 2014; 40: 772–784.
Sakuma K, Aoki M, Kannagi R . Transcription factors c-Myc and CDX2 mediate E-selectin ligand expression in colon cancer cells undergoing EGF/bFGF-induced epithelial-mesenchymal transition. Proc Natl Acad Sci USA 2012; 109: 7776–7781.
Jiang H, Fu XG, Chen YT . Serum level of endothelial cell-specific molecule-1 and prognosis of colorectal cancer. Genet Mol Res 2015; 14: 5519–5526.
Na YJ, Kim YH, Ye JJ, Yun HK, Lee CI . Identification of endothelial cell-specific molecule-1 as a potential serum marker for colorectal cancer. Cancer Sci 2010; 101: 2248–2253.
Kang UB, Yeom J, Kim HJ, Kim H, Lee C . Expression profiling of more than 3500 proteins of MSS-type colorectal cancer by stable isotope labeling and mass spectrometry. J Proteomics 2012; 75: 3050–3062.
Song M, Kim H, Kim WK, Hong SP, Lee C, Kim H . High expression of AT-rich interactive domain 3A (ARID3A) is associated with good prognosis in colorectal carcinoma. Ann Surg Oncol 2014; 21: 481–489.
Covarrubias AS, Bergfors T, Jones TA, Högbom M . Structural mechanics of the pH-dependent activity of beta-carbonic anhydrase from Mycobacterium tuberculosis. J Biol Chem 2006; 281: 4993–4999.
Kummola L, Hämäläinen JM, Kivelä J, Kivelä AJ, Saarnio J, Karttunen T et al. Expression of a novel carbonic anhydrase, CA XIII, in normal and neoplastic colorectal mucosa. BMC Cancer 2005; 5: 41.
Parri M, Pietrovito L, Grandi A, Campagnoli S, Camilli ED, Bianchini F et al. Angiopoietin-like 7, a novel pro-angiogenetic factor over-expressed in cancer. Angiogenesis 2014; 17: 881–896.
Moutinho C, Martinez-Cardús A, Santos C, Navarro-Pérez V, Martínez-Balibrea E, Musulen E et al. Epigenetic inactivation of the BRCA1 interactor SRBC and resistance to oxaliplatin in colorectal cancer. J Natl Cancer Inst 2013; 106: 91–103.
Sopik V, Phelan C, Cybulski C, Narod SA . BRCA1 and BRCA2 mutations and the risk for colorectal cancer. Clin Genet 2015; 87: 411–418.
Niell BL, Rennert G, Bonner JD, Almog R, Tomsho LP, Gruber SB . BRCA1 and BRCA2 founder mutations and the risk of colorectal cancer. J Natl Cancer Inst 2004; 96: 15–21.
Finlin BS, Gau CL, Murphy GA, Chiu YF, Botstein D, Brown PO et al. RERG Is a novel ras-related, estrogen-regulated and Growth-inhibitory Gene in Breast Cancer. J Biol Chem 2001; 276: 42259–42267.
Mitchell SM, Ross JP, Drew HR, Ho T, Brown GS, Saunders NF et al. A panel of genes methylated with high frequency in colorectal cancer. BMC Cancer 2014; 14: 740–745.
Zou H, Harrington JJ, Shire AM, Rego RL, Liang W, Campbell ME et al. Highly methylated genes in colorectal neoplasia: implications for screening. Cancer Epidemiol Biomark Prev 2007; 16: 2686–2696.
Lin K, Taylor JR, Wu TD, Gutierrez J, Elliott JM, Vernes JM et al. TMEFF2 is a PDGF-AA binding protein with methylation-associated gene silencing in multiple cancer types including glioma. PLoS ONE 2011; 6: e18608–e18608.
Sun TT, Tang JY, Du W, Zhao HJ, Zhao G, Yang SL et al. Bidirectional regulation between TMEFF2 and STAT3 may contribute to H. pylori-associated gastric carcinogenesis. Int J Cancer 2014; 136: 1053–1064.
Barsky SH, Siegal GP, Jannotta F, Liotta LA . Loss of basement membrane components by invasive tumors but not by their benign counterparts. Lab Invest 1983; 49: 140–147.
Thiery JP . Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2002; 2: 442–454.
Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T . Opinion: migrating cancer stem cells—an integrated concept of malignant tumour progression. Nat Rev Cancer 2005; 5: 744–749.
Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology 2006; 131: 830–840.
Xiong H, Hong J, Du W, Lin Y, Ren L, Wang Y et al. Roles of STAT3 and ZEB1 proteins in E-cadherin down-regulation and human colorectal cancer epithelial-mesenchymal transition. J Biol Chem 2011; 287: 5819–5832.
Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res 2008; 68: 537–544.
Zhang G, Zhou T, Tian H, Liu Z, Xia S . High expression of ZEB1 correlates with liver metastasis and poor prognosis in colorectal cancer. Oncol Lett 2013; 5: 564–568.
Crous-Bou M, Rennert G, Salazar R, Rodriguez-Moranta F, Rennert HS, Lejbkowicz F et al. Genetic polymorphisms in fatty acid metabolism genes and colorectal cancer. Mutagenesis 2012; 27: 169–176.
Yeh CS, Wang JY, Cheng TL, Juan CH, Wu CH, Lin SR . Fatty acid metabolism pathway play an important role in carcinogenesis of human colorectal cancers by microarray-bioinformatics analysis. Cancer Lett 2006; 233: 297–308.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Cancer Gene Therapy website
Supplementary information
Rights and permissions
About this article
Cite this article
Liu, H., Zhang, C. Identification of differentially expressed genes and their upstream regulators in colorectal cancer. Cancer Gene Ther 24, 244–250 (2017). https://doi.org/10.1038/cgt.2017.8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cgt.2017.8
This article is cited by
-
Activation of CHPF by transcription factor NFIC promotes NLRP3 activation during the progression of colorectal cancer
Functional & Integrative Genomics (2024)
-
Panomics reveals patient individuality as the major driver of colorectal cancer progression
Journal of Translational Medicine (2023)
-
Nuclear factor I-C overexpression promotes monocytic development and cell survival in acute myeloid leukemia
Leukemia (2023)
-
Integrated nuclear proteomics and transcriptomics identifies S100A4 as a therapeutic target in acute myeloid leukemia
Leukemia (2020)
-
Identification of Pathogenic Genes and Transcription Factors in Osteosarcoma
Pathology & Oncology Research (2020)