Abstract
How metabolic pathways required for epidermal tissue growth and remodeling influence the ability of keratinocytes to survive stressful conditions is still largely unknown. The mechanistic target of rapamycin complex 2 (mTORC2) regulates growth and metabolism of several tissues, but its functions in epidermal cells are poorly defined. Rictor is an adaptor protein essential for mTORC2 activity. To explore the roles of mTORC2 in the epidermis, we have conditionally deleted rictor in mice via K14-Cre-mediated homologous recombination and found that its deficiency causes moderate tissue hypoplasia, reduced keratinocyte proliferation and attenuated hyperplastic response to TPA. Noteworthy, rictor-deficient keratinocytes displayed increased lifespan, protection from senescence, and enhanced tolerance to cellular stressors such as growth factors deprivation, epirubicin and X-ray in vitro and radioresistance in vivo. Rictor-deficient keratinocytes exhibited changes in global gene expression profiles consistent with metabolic alterations and enhanced stress tolerance, a shift in cell catabolic processes from glycids and lipids to glutamine consumption and increased production of mitochondrial reactive oxygen species (ROS). Mechanistically, the resiliency of rictor-deficient epidermal cells relies on these ROS increases, indicating stress resistance via mitohormesis. Thus, our findings reveal a new link between metabolic changes and stress adaptation of keratinocytes centered on mTORC2 activity, with potential implications in skin aging and therapeutic resistance of epithelial tumors.
Similar content being viewed by others
Main
Adaptation to stressful conditions is key for organisms evolution and the epidermis contributes to this function providing a barrier against physical and chemical injuries, dehydration and pathogens infection.1
From yeast to mammals, stress tolerance and lifespan extension are favored by low-nutrient conditions leading to increased metabolic rates.2, 3 Caloric restriction, low glucose intake and inhibition of the insulin/IGF pathway parallel with extended lifespan and mild increases of reactive oxygen species (ROS).4, 5 Albeit traditionally considered as purely harmful, ROS are physiological regulators of stress-response mechanisms that prevent cellular damage (refs 3, 6 and references therein). Mitohormesis is a process initiated by moderate increases of mitochondrial ROS, which enhance resistance to stressors by engaging programs of cell protection (ref. 3 and references therein). Although molecular pathways including Akt, p53, Nrf2, NFκB and AP1 regulate the epidermal responses to oxidative damage, UV and X-ray radiations and chemotherapic drugs,7, 8, 9 whether mitohormesis has a role in keratinocyte stress adaptation is unknown.
The mechanistic target of rapamycin (mTOR), an evolutionarily conserved serine/threonine protein kinase, is a signaling hub integrating cell growth, metabolism and energy stress responses.10 mTOR signaling is activated by extracellular cues such as nutrients and growth factors, and favors cellular and organismal growth by stimulating biosynthesis of macromolecules. mTOR inhibition causes lifespan extension via mitohormesis in yeast,11 and prevents energy expenditure by promoting autophagic recycling of cellular components.12 mTOR kinase is the catalytic subunit of two signaling complexes, mTOR Complex1 (mTORC1) and mTOR Complex 2 (mTORC2).13 Although most mTOR biological functions have been attributed to mTORC1,14 mTORC2 regulates AGC family protein kinases such as Akt and PKC isoforms necessarily adjuvated by the evolutionarily conserved adaptor protein Rictor.13 By phosphorylating Akt proteins at a regulatory residue (Ser473 in Akt1) in response to growth factors, mTORC2 promotes maximal Akt kinase activity and cell survival in mammals, and regulates actin cytoskeleton via PKC signaling.15, 16
In mammals, germline ablation of rictor is incompatible with development.17 Although tissue-specific embryonic ablation of rictor interferes with morphogenesis in the vascular endothelium and nervous systems,18, 19 its deletion in skeletal muscle, adipose tissue and liver impairs lipogenesis and glucose metabolism.20, 21, 22
Here we show that rictor conditional ablation during epidermal development causes tissue hypoplasia in the newborn mice paralleled with attenuated keratinocyte proliferation rates. Importantly, rictor/mTORC2 deficiency in keratinocytes causes delayed senescence, enhanced resistance to cellular stressors, and a shift of catabolic functions towards glutaminolysis, elevated mitochondrial activity and ROS production. These ROS increases have a mitohormetic role, being crucial for the resiliency of rictor-deficient keratinocytes. Thus, our work implicates rictor/mTORC2 as a novel signaling node integrating epidermal metabolism with stress adaptation.
Results
Rictor/mTORC2 disruption in murine epidermis leads to tissue hypoplasia in newborn mice
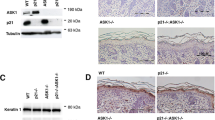
To conditionally delete rictor in the epidermis, mice carrying a rictor allele in which exon 3 is flanked by two loxP sites17 were bred with mice expressing Cre recombinase under the control of the keratin 14 (K14) promoter23 (Figure 1a). Newborn mice with K14-Cre-mediated homozygous deletion of rictor (E-RiKO mice) displayed undetectable levels of rictor protein in the epidermis while the mTORC1-specific raptor levels were unchanged (Figure 1b) compared with control (CT) mice. E-RiKO mice were born at the expected Mendelian rate and did not show obvious epidermal or hair follicle abnormalities from birth until 1 year of age (not shown). Immunoblotting analysis confirmed ablation of rictor in E-RiKO keratinocytes paralleled by nearly abrogated phosphorylation of Ser473 Akt in response to growth factors, whereas phosphorylation of Thr308 Akt was preserved. Keratinocytes, like other cell types, exhibited reduced total Akt and PKCα levels following rictor ablation (Figure 1c)13 without significant alterations of mTORC1 signaling or Akt targets phosphorylation, except for attenuated phosphorylation of FoxO1 and -3 (ref. 24 and Supplementary Figure S1a), thereby displaying features typical of mTORC2 disruption.
Loss of rictor/mTORC2 in the epidermis results in tissue hypoplasia and impaired TPA response. (a) Diagram of the breeding strategy used to obtain K14-cre-mediated homozygous deletion of rictor (E-RiKO). WT: wild-type mouse, CT: WT mouse with exon 3 flanked by two LoxP sites. White tile: hexon; Black arrowhead: LoxP site. (b and c) Representative western blotting analysis with the indicated antibodies of (b) epidermal extracts of CT and E-RiKO newborn (P3) littermates (n=3); (c) cell extracts of CT and E-RiKO MPKs subjected to starvation/stimulation treatment for the indicated times. (d and e) Skins of E-RiKO and CT littermates (P3): (d) representative H&E staining (left panels) and histograms of the epidermal thickness; Epi: epidermis, Der: dermis; scale bar: 30 μm; (e) PCNA staining (left panels) and histograms of the percentage of PCNA+ epidermal area. Arrows: brightest PCNA+ cells in epidermal basal layers; scale bar: 50 μm. (f) Representative H&E images of skin section obtained from E-RiKO and CT littermates (7-week old) on 48 h of TPA or vehicle treatment, (n=3); scale bar: 30 μm. Histograms represent epidermal thickness. (g) Representative confocal images of IF performed on skin sections, obtained from mice treated as in f and injected with BrdU for the final hour of the experiment, with anti-BrdU antibody (red) and counterstained with anti-LaminA antibody (green). Arrowheads: BrdU+ cells; scale bar: 50 μm. The percentage of BrdU+ cells/area (μm2) was quantified (right histograms). Quantification of epidermal thickness (mean±SEM) for three mice/genotype; the standard error and the significance of the differences between groups were determined with linear regression using GEE. ***P<0.0005. PCNA and BrdU histograms represent the mean±S.D. of 30 fields/genotype (n=3). ***P<0.0005
Compared with CTs, E-RiKO epidermis was stratified but thinner (Figure 1d) in newborn animals. Such hypoplasia was unlikely the result of cell death as we did not detect TUNEL- or cleaved caspase-3 positivity in the epidermis (not shown). In contrast, positivity for the PCNA proliferative marker was reduced in E-RiKO mice (Figure 1e). p63 and loricrin, used as readouts of progenitor and differentiated keratinocytes, respectively, were expressed with proper spatial tissue distribution albeit at lower levels in mutant mice (Supplementary Figures S1b and c), suggesting that hypoplasia reflects impaired cell growth affecting the cellularity of different epidermal compartments. Instead, the thickness of adult murine epidermis, composed by only two to three cellular layers, was comparable between CT and mutant mice (Figure 1f). TPA treatment of adult epidermis induces rapid keratinocyte proliferation resulting in hyperplasia by 48 h.25 Seven-week-old E-RiKO mice displayed attenuated epidermal thickening upon TPA exposure (Figure 1f), paralleled by a decreased BrdU incorporation (Figure 1g). Thus, mTORC2 deficiency restrains growth and hyperplasia of the epidermis in part by attenuating mitogenic responses.
Rictor/mTORC2 deficiency impairs keratinocytes proliferation and delays senescence in vitro
We compared primary keratinocytes derived from CT or E-RiKO newborn littermates under proliferating conditions (low calcium medium; LCM) and monitored cultures until CT cells reached senescence. Consistent with the hypoplastic phenotype, the number of keratinocytes isolated from E-RiKO epidermis was reduced relative to CT counterparts (Figure 2a). E-RiKO keratinocytes displayed attenuated growth rates (Figure 2b), decreased percentages of BrdU+ cells (Figure 2c and Supplementary Figure S2a), reduced cell density (Figure 2d), decreased S-phase and a proportional increase in the G0/G1 phase (Supplementary Figure S2b). Notably, whereas CT keratinocytes acquired a senescent-like morphology, E-RiKO cells exhibited proliferative appearance until at least 20–25 days from plating (Figure 2d) and lower levels of p16, p19 and p53 senescence markers (Figure 2e) and β-galactosidase activity (Figure 2f). Moreover, albeit LCM culture conditions are unfavorable for spontaneous keratinocyte immortalization,26 E-RiKO cultures became immortalized with 100% efficiency as compared with ~18% of CTs (Supplementary Figures S2c–e).
Rictor-deficient keratinocytes display reduced proliferation and delayed senescence in vitro. (a) Quantification of MPKs derived from at least 10 skins of CT and E-RiKO newborn mice (P3). Histograms represent mean±S.D. ***P<0.0005. E-RiKO and CT MPKs were isolated from littermates P3 (n=5) and analyzed as follows. (b) Growth curve: MPKs (both adherent and suspended/differentiated cells) were detached and counted at the indicated times upon plating. Data are mean±S.D. obtained from triplicate samples/genotype of three independent experiments. CompareGrowthCurve function was used (see the ‘Materials and Methods’ section); P=0.029. (c) Confluent MPKs, were subjected to a single BrdU pulse (3 h) under basal conditions. Histograms represent mean±S.D. of the percentage of BrdU+ cells determined from at least 200 cells/genotype of three independent experiments; ***P<0.0005. (d) Representative phase contrast images of MPKs cultured in LCM for the indicated times. Note that E-RiKO cells show at 3 days reduced density compared with CT counterparts; at 5–10 days, they display a comparable confluency; while at 25 days, they maintained an undifferentiated proliferative morphology; whereas CT cells show a flattened, senescent appearance; scale bar: 100 μm. (e) Western blotting analysis for the indicated markers of MPKs extracts of 25-day cultures. (f) Representative images of 25-day-old MPKs stained for SA-βGal (left panel). The percentage of SA-βGal+ cells is quantified in the histograms (right) as mean±S.D. of at least 200 cells/genotype of three independent experiments; scale bar: 50 μm; *P<0.05
Rictor-deficient keratinocytes are protected from death induced by multiple cellular stressors
Several molecular determinants of cell senescence are also implicated in cell death. Upon growth factors deprivation, E-RiKO cells displayed reduced morphological death signs (Supplementary Figure S3a), delayed/attenuated caspase-3 cleavage (Cl-Casp3; Figure 3a) and decreased cell death by AnnexinV/DAPI analysis (Figure 3b). By comparing the responses to epirubicin, an anticancer drug that induces cell death via DNA damage and oxidative stress,27 the majority of CT cells detached after 15 h of treatment, whereas many E-RiKO cells seemed unaffected (Supplementary Figure S3a) and displayed lower Cl-casp3 levels (Figure 3c) and reduced death (Figure 3d). As the S-phase of the cell cycle renders cells vulnerable to epirubicin, we verified whether the reduced sensitivity of E-RiKO keratinocytes reflected their reduced division rates by exposing cells to the drug under comparable growth arrested conditions (Figure 3e). Even in this case, E-RiKO cells displayed lower Cl-casp3 levels (Figure 3f), suggesting that their resiliency does not simply depend on attenuated proliferation.
Rictor-deficient keratinocytes display resistance to death-inducing stimuli both in vitro and in vivo. (a–h) E-RiKO and CT MPKs (P3) were analyzed at confluency (5–6 days in culture) as follows. (a) Representative western blotting analysis of extracts obtained from E-RiKO and CT MPKs on starvation (st) for the apoptosis marker cleaved caspase-3 (Cl-casp3) and Rictor. Loading control: Tubulin. (b) Representative AnnexinV/DAPI flow cytometry plots of E-RiKO and CT MPKs kept in complete LCM (−) or starvation (St) for 72 h. Histograms are mean±S.D. of dead cells expressed as variation of the fraction of dead cells in response to starvation (72 h) (see the ‘Materials and Methods’ section) *P<0.05. (c and d) MPKs grown in LCM for 24 h were kept untreated (−) or treated with epirubicin (10 μM) for 18 h: (c) Representative Western blotting analysis for the indicated proteins. (d) Representative AnnexinV/DAPI flow cytometry plots of E-RiKO and CT MPKs kept untreated (−) or treated with Epirubicin for 15 h. Histograms are mean±S.D. of dead cells determined as in b; *P<0.05. (e) MPKs maintained in LCM or starved (St) for 18 h were treated with a single BrdU pulse. BrdU+ cells were determined out of 200 cells (DAPI+)/genotype in three independent experiments. Histograms represent mean±S.D. of BrdU+ cells. Note that on St 18 h, cells display comparable BrdU uptake; *P<0.05; ***P<0.0005. (f) MPKs were starved as in e and then treated with the indicated doses of epirubicin for 4 h. Cell extracts were analyzed by western blotting for Rictor, Cl-casp3 and Tubulin, loading control. Ratio between Cl-casp3 and Tubulin is reported. (g) Histograms represent mean colony number±S.D. from duplicate plates determined in untreated (−) and treated (X-ray) MPKs colony-forming efficiency assay, relative to the value obtained with CT (−) cells set as unitary. Data are representative of at least two independent experiments; ***P<0.0005, **P<0.005. (h) Representative AnnexinV/DAPI flow cytometry plots of E-RiKO and CT MPKs kept untreated (−) or X-ray treated (60 Gy) analyzed after 96 h. Histograms are mean±S.D. of dead cells determined as in b; *P<0.05. (i) Representative confocal images for TUNEL (red) and DAPI (blue) stainings of skin sections obtained from E-RiKO and CT littermates (P3) subjected to a full-body single dose of X-ray radiation (8 Gy) and sacrificed 24 h later; scale bar: 100 μm; n=5 mice/genotype. (j) Quantification of TUNEL staining of X-ray-treated skins with 4 Gy and 8 Gy as in i. Histograms represent mean±S.D. of TUNEL+ cells/area (103 μm2) of at least 30 hair follicles of four mice/genotype. Note that the TUNEL+ area was limited to the hair follicle matrix, with similar cellular density and sensitive to radiations; ***P<0.0005
We next evaluated the effects of different X-ray doses. E-RiKO cells retained higher clonogenic ability upon X-ray exposure (Figure 3g), confirming enhanced stress tolerance. Keratinocytes are highly refractory to X-ray-induced apoptosis;28 hence, we evaluated cell death by AnnexinV/DAPI analysis after one 60 Gy dose, which induces ~30% of cell death in CT keratinocytes. E-RiKO cells had a ~25% reduction in cell death (Figure 3h). Irradiated E-RiKO cells showed attenuated Cl-casp3 (Supplementary Figure S3d), and displayed a reduction in both senescence (Supplementary Figures S3e and S3f) and growth arrest in response to X-ray (Supplementary Figure S3g). Mutant keratinocytes exhibited reduced H2AX phosphorylation (γH2AX) suggestive of lower DNA damage (Supplementary Figure S3d), and decreased Chk1 and Chk2 levels (Supplementary Figure S3d) similarly to cancer cells subjected to rictor or mTOR ablation.29 Interestingly, in CT cells, Ser473 Akt phosphorylation decreased early on after X-ray exposure. In contrast, treatment of E-RiKO and CT keratinocytes with the DNA-damaging agent cisplatin did not reveal significant differences in morphology, sensitivity to apoptosis, levels of Cl-casp3 and γH2AX, and in CT cells Akt Ser473 phosphorylation was not reduced at early times (Supplementary Figures S3a–c). Thus, E-RiKO keratinocytes are not resistant to death per se and retain functional apoptotic machinery.
To verify whether rictor deficiency protects keratinocytes from X-ray-induced cell death in vivo, we analyzed the skins of irradiated E-RiKO and CT littermates by TUNEL assay. Consistent with previous reports, positivity to the staining was minimal in the interfollicular epidermis,30 and TUNEL+ keratinocytes were confined within the hair follicle matrix31 (Figure 3i). Notably, E-RiKO mice displayed a significant decrease in TUNEL+ cells compared with CT littermates (Figure 3j).
Gene expression profiles of rictor-deficient keratinocytes indicate metabolic alterations under basal conditions and upon X-ray exposure
To identify specific genes and pathways involved in the enhanced stress tolerance of E-RiKO keratinocytes, we performed a RNA-seq transcriptome analysis by comparing mutant and CT cells under basal growing conditions, and at different times after X-ray exposure (Supplementary Table S1). Under basal conditions, 589 genes were differentially regulated between E-RiKO and CT cells, that is, 336 downregulated and 253 upregulated genes in the former (Figure 4a, Supplementary Tables S2 and S3). Gene Ontology (GO) enrichments were considered significant with a nominal P-value less than 10−3 with Fisher’s exact test. Rictor deficiency was associated with significant enrichment of genes involved in lipid metabolism, keratinocyte differentiation, oxidation–reduction process, lipid catabolic process and lipid biosynthetic process among downregulated genes, and genes involved in cell motility, signal transduction, inflammatory response, response to stress and defense response among upregulated genes (Figure 4b). The complete GO analysis is provided in Supplementary Tables S4 and S5.
RNA-Seq analysis of rictor-deficient keratinocytes suggests a link between metabolic changes and stress adaptation. E-RiKO and CT MPKs were isolated from newborn littermates (n=6/genotype), grown in LCM and analyzed by RNA-Seq analysis under basal conditions (24 h LCM) and upon X-ray exposure (8 Gy) for 1 h and 24 h. (a) Upregulated and downregulated genes detected in E-RiKO versus CT cells under basal conditions. (b) Selected gene ontology (GO) categories enriched in differentially expressed genes as in a. The number of genes belonging to each GO category is indicated. Rictor deficiency was coupled with downregulation of genes involved in lipid metabolism (P=1.08E−16), keratinocyte differentiation (P=1.68E−9), oxidation–reduction process (P=1.89E−6), lipid catabolic process (P=2.41E−6), lipid biosynthetic process (P=2.7E−6) and upregulation of genes involved in cell motility (P=2.62E−18), signal transduction (P=3.15E−15), inflammatory response (P=1.66E−12), response to stress (P=3.11E−7) and defense response (P=2.28E−6) as in b. (c) Upregulated and downregulated genes of X-ray treated MPKs of the indicated genotype relative to basal conditions. (d) Differentially regulated genes overlap between CT MPKs X-ray treated for 1 h and E-RiKO MPKs under basal conditions (27.5% gene overlap; P=5.44E−57, Fisher’s exact test). (e) Selected GO categories enriched in overlapping differentially regulated genes (DEGs) subgroup as in d
By evaluating the number of modulated genes after 1 h of X-ray exposure versus basal conditions, E-RiKO keratinocytes displayed a dramatically reduced response to the treatment (i.e., 94 versus 794 genes), while after 24 h the differences between genotypes were attenuated (Figure 4c).
Based both on these results and the greater stress tolerance of mutant cells, we hypothesized that genes modulated in CT cells following treatment might be similarly up- or downregulated in E-RiKO under basal conditions. To this aim, we compared differentially expressed genes (DEGs) in CT cells under X-ray treated (1 h) versus basal conditions, with DEGs in E-RiKO versus CT cells under basal conditions (Figure 4d). Notably, we found a highly significant 27.5% genes overlap (P=5.44E−57), corresponding to 161 DEGs (62% downregulated and 38% upregulated). The majority of these genes were expressed at similar levels in basal E-RiKO cells and X-ray exposed CT cells (Supplementary Tables S1). GO analysis of this subgroup of genes revealed an enrichment in lipid metabolic process, lipid catabolic process, response to hypoxia and lipid biosynthetic process in the downregulated class whereas few GO survived in the upregulated class at this stringency, among which regulation of epithelial cell proliferation and response to glucose (Figure 4e, Supplementary Tables S6–7). These data suggest that rictor deficiency alters the metabolic functions of keratinocytes under basal conditions, and because several metabolic genes that were rapidly turned down in CT cells in response to X-ray were basally downregulated in E-RiKO cells, this suggests a link between metabolic changes and stress adaptation.
Rictor deficiency in keratinocytes promotes metabolic rewiring and ROS production
ROS contribute to cell death mechanisms triggered by growth factors deprivation, epirubicin and X-ray, while cisplatin, to which E-RiKO cells are sensitive, promotes cell death primarily via direct DNA damage independently of ROS.32 To our surprise, we detected higher basal levels of ROS (~1.7-fold) in E-RiKO keratinocytes relative to CT counterparts (Figure 5a), while 24 h after X-ray exposure CT cells displayed a more robust increase (~2.9-fold versus 1.2-fold; Supplementary Figure S3f). Mitochondrial ROS were also higher in E-RiKO cells (Figures 5b and c). By contrast, the activity of aldose reductase, a source of cytosolic ROS,33 was reduced of 20% in E-RiKO (Figure 5d) while NADPH oxidase activity, which also generates cytosolic ROS, was similar (Figure 5e). The electron transport chain (Figure 5f) and the amounts of mitochondrial ATP (Figure 5g) were instead increased, suggesting that rictor deficiency promotes increases of mitochondrial ROS and respiration.
Rictor-deficient keratinocytes display metabolic reprogramming. E-RiKO and CT MPKs, isolated and cultured in LCM (n=5 littermates P3/genotype), were analyzed at confluency in LCM 24 h as follows: (a) total cellular ROS (nmol/mg prot); (b) mitochondrial ROS (nmol/mg mit prot); (c) mitochondrial/total ROS (%) determined by compairing mitochondrial and total ROS, each normalized for total protein content; (d) aldose reductase (nmol NADP+/min/mg prot); (e) NADPH oxidase (RLU/mg prot); (f) electron transport chain (nmol red cit c/min/mg mit prot); (g) ATP (nmol/mg mit prot); (h) lipid β-oxidation (pmol/h/mg prot); (i) triglycerides (nmol/mg prot); (j) glucose uptake (pmol glucose/mg prot); (k) hexokinase (nmol NADH/min/mg prot); (l) phosphofructose kinase 1 (nmol NAD+/min/mg prot); (m) glutaminase (μmol NADH/min/mg prot); (n) glutamic dehydrogenase (μmol NADH/min/mg prot); (o) TCA cycle with [14C] glutamine (pmol CO2/h/mg mit prot); (p) TCA cycle with [14C] glutamate (pmol CO2/h/mg mit prot); (q) intracellular [14C]glutamine (μmol/mg prot); (r) intracellular [14C]glutamate (μmol/mg prot); (s) glutamate/glutamate ratio; (t) total SOD (μmol/min/mg prot); (u) catalase (nmol/min/mg prot); (v) mitochondrial SOD (μmol/min/mg prot). All histograms represent mean±S.D. of the indicated metabolic parameters determined in at least three independent experiments; *P<0.05; **P<0.005; ***P<0.0005
Consistent with mTORC2 deficiency being coupled with defective lipogenesis,21, 22 E-RiKO cells had reduced triglycerides levels (Figure 5i), as suggested by GO analysis. We then analyzed the activity of catabolic pathways that may impinge on ROS production by fueling mitochondrial oxidative phosphorylation. Fatty acid β-oxidation was reduced to ~60% of CT values in E-RiKO cells (Figure 5h), in agreement with RNA-seq data (Figure 4b). Moreover, E-RiKO cells showed decreased glucose uptake and glycolysis (Figures 5j–l). The glutaminolysis energetic pathway provides carbon source alternative to lipids and glycids, nitrogen for nucleotide biosynthesis and NADPH for redox maintenance.34 E-RiKO keratinocytes showed increased activity of both glutaminase (2.3-fold) and glutamic dehydrogenase (1.5-fold), which catalyze the first and the second step of glutaminolysis, respectively, although we did not detect alterations in their mRNA and/or protein levels (Figures 5m and n, Supplementary Table S1 and data not shown). Moreover, in both l-[14C]-glutamine- or l-[14C]-glutamate-labeled cells, the flux through the tricarboxylic acid (TCA) cycle was higher in rictor-deficient keratinocytes, and was reduced by the glutaminase inhibitor BPTES ((1Z,1’Z)-N’,N’’-(5,5’-(thiobis(ethane-2,1-diyil))bis(1,3,4-thiadiazole-5,2-diyil))bis(2-phenylacetimidic acid); Supplementary Figure S4a), in l-[14C]-glutamine-treated cells but not in l-[14C]-glutamate-labeled cells (Figures 5o and p), suggesting that TCA cycle was strongly fueled by the glutaminolytic anaplerotic reaction in E-RiKO cells.
As the production of glutamate represents the first obligatory step in glutamine catabolism, the ratio between intracellular l-glutamate versus l-glutamine – after pulsing cells with l-[14C]-glutamine – was used as readout of glutamine consumption: although glutamine uptake was similar between genotypes, in E-RiKO cells both the production of glutamate and the ratio between l-glutamate and l-glutamine was more elevated (Figures 5q–s). Thus, rictor deficiency promotes keratinocyte metabolic reprogramming, diverting the catabolism from lipids and glycids to glutamine consumption.
To verify whether E-RiKO cells possessed higher antioxidant capacity, we measured the activities of the main antioxidant enzymes, cytosolic and mitochondrial superoxide dismutases (SODs) and catalase. Mitochondrial SOD activity was more than doubled in E-RiKO cells compared with CTs (Figures 5t–v), whereas cytoplasmic SOD and catalase activities did not differ significantly. Because the levels of catalase, SOD1 and SOD2 proteins, which account for SOD activities in cytosol and mitochondria, respectively, were unchanged (Supplementary Figure S4b, Supplementary Table S1 and data not shown), the enhanced mitochondrial SOD activity of E-RiKO cells may rely on post-translational modifications of SOD2.
Rictor-deficient keratinocytes are sensitized to epirubicin-induced cell death by antioxidant treatment and glutaminase inhibition
To define whether the increased ROS of E-RiKO cells are responsible for their stress-resistant phenotype, we treated E-RiKO and CT keratinocytes with epirubicin in the presence of ROS scavenger N-acetyl cysteine (NAC) at a concentration able to lower total and mitochondrial ROS in E-RiKO cells to levels close to untreated CT cells (Figures 6a and b). Without NAC, E-RiKO keratinocytes resulted more resistant to apoptosis in response to epirubicin, whereas NAC-treated E-RiKO keratinocytes displayed Cl-casp3 and AnnexinV/DAPI profiles close to those of CT cells unexposed to NAC (Figures 6c and d), indicating that this compound sensitizes E-RiKO cells to epirubicin-induced apoptosis. This was apparently in contrast to findings reporting that cells are protected from epirubicin-induced death by a NAC pretreatment up to 24 h.35 Under these conditions, we could detect a significant reduction of Cl-casp3 (Supplementary Figure S5a) in CT keratinocytes, but we were unable to restore normal ROS levels in E-RiKO cells (Supplementary Figure S5b). These data suggest that prolonged versus transient ROS scavenging has different effects on the stress tolerance of normal keratinocytes. Also Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid), a distinct antioxidant compound, restored normal ROS levels in E-RiKO cells and increased Cl-casp3 levels in response to epirubicin (Supplementary Figure S5c and d).
ROS scavenging and glutaminase inhibition sensitize rictor-deficient keratinocytes to epirubicin-induced cell death. E-RiKO and CT MPKs were isolated (n=5/genotype), grown in LCM and analyzed at confluency for the following. (a) Total cellular ROS was measured on LCM (−) or NAC 10 mM for 48 h. Histograms represent mean±S.D. of ROS levels measured in at least three independent experiments; ***P<0.0005; *P<0.05. (b) Histograms represent mean±S.D. of mitochondrial ROS determined in cells treated as in a. (c) Representative western blotting analysis for the indicated antibodies of cell extracts obtained from MPKs maintained as in a and subsequently treated with DMSO (−) or epirubicin 10 μM for 10 h. Ratio between Cl-casp3 and keratin5 (K5) is reported. (d) Representative AnnexinV/DAPI flow cytometry plots of E-RiKO and CT MPKs maintained untreated (−) or pre-treated with NAC and subsequently treated with epirubicin 10 μM for 15 h. Histograms are mean±S.D. of dead cells determined as described in the ‘Materials and Methods’ section; *P<0.05. (e) Histograms represent the mean±S.D. of the glutaminase activity (μmol NADH/min/mg prot) evaluated in CT and E-RiKO MPKs treated with BPTES (10 μM, 2 h) in at least three independent experiments; **P<0.005, *P<0.05. (f) Histograms represent mean±S.D. of total ROS levels (nmol/mg prot) measured in cells treated as in e in at least three independent experiments; *P<0.05. (g) Representative western blotting analysis with the indicated antibodies of cell extracts derived from E-RiKO and CT cells pre-treated with BPTES at the indicated doses for 48 h and exposed to epirubicin in the presence of BPTES for 15 h. Ratio between Cl-casp3 and keratin5 (K5) is reported. (h) Representative AnnexinV/DAPI flow cytometry plots of E-RiKO and CT MPKs maintained untreated (−) or pre-treated with BPTES and subsequently treated with epirubicin 10 μM for 15 h. Histograms are mean±S.D. of dead cells as in d; **P<0.005, *P<0.05
NAC exposure of CT and E-RiKO cells did not affect other metabolic parameters (Supplementary Figure S5e), apart from the decrease of mitochondrial SOD activity in mutant cells to levels similar to untreated CT cells.
These data suggest that the activity of NAC is primarily due to its antioxidant capacity, and that the increased ROS are not the inducers of the metabolic changes produced by rictor deficiency but rather their consequence.
The increased ROS of E-RiKO cells may depend on increased glutaminolytic activity. Consistent with this hypothesis, BPTES effectively reduced glutaminase activity in both genotypes and restored in E-RiKO-treated cells ROS levels similar to CT cells (Figures 6e and f), indicating glutaminase as a major determinant of ROS increases. Importantly, sustained glutaminase inhibition also phenocopied the effects of NAC in E-RiKO cells, as it increased both Cl-casp3 levels and cell death in response to epirubicin (Figures 6g and h). Thus, both the re-establishment of normal ROS levels and the inhibition of their principal metabolic source sensitize E-RiKO cells to stressors to which they are typically resistant. ROS scavenging by NAC also attenuated features of E-RiKO cells beyond stress response such as their decreased BrdU uptake (Supplementary Figure S5f) and protection from senescence (Supplementary Figure S5g), suggesting the involvement of ROS also in these phenotypes.
The epidermal metabolic, molecular and stress-response phenotypes of E-RiKO mice indicate a critical role of ROS in stress protection
To determine whether metabolic alterations observed in cultured E-RiKO keratinocytes were present in the epidermis of mutant mice in vivo, we compared several metabolic parameters in fresh epidermal extracts obtained from CT and E-RiKO mice (Figure 7a). Relative to CT mice, total and mitochondrial ROS, electron transport chain activity, ATP content, glutaminase and glutamic dehydrogenase activities were all more elevated in mutant mice, while lipid β-oxidation and hexokinase activities were reduced, reflecting closely the findings on cultured cells.
Rictor-deficient epidermis displays a metabolic rewiring and ROS-dependent lower sensitivity to stress. (a) Metabolic analysis of CT and E-RiKO skins obtained from at least three mice/genotype. Histograms represent mean±S.D. of the following metabolic parameters: total cellular ROS (nmol/mg prot); mitochondrial ROS (nmol/mg mit prot); electron transport chain (nmol red cit c/min/mg mit prot); ATP (nmol/mg mit prot); lipid β-oxidation (pmol/h/mg prot); hexokinase (nmol NADH/min/mg prot); glutaminase (μmol NADH/min/mg prot); glutamic dehydrogenase (μmol NADH/min/mg prot); **P<0.005, *P<0.05. (b) RNA was extracted from CT and E-RiKO epidermis of at least six mice/genotype and RT-PCR analysis for the following genes was performed: Gadd45-α; Gadd45-β; Gadd45-γ; Glul; Il1-α; Acsl1; Faah; Fabp4. Data are represented as mean±S.D. of the fold change relative to unitary value assigned to CT; **P<0.005, *P<0.05 (c) Representative confocal images of IF for TUNEL (red) and DAPI (blue) stainings on E-RiKO and CT skin sections pre-treated with NAC (or vehicle) for 48 h (i.p. injection), subjected to a single dose of X-ray radiation (8 Gy) and collected 24 h later; scale bar: 100 μm; n=10 mice/genotype. (d) Histograms represent mean±S.D. of ROS levels (nmol/mg prot) in CT and E-RiKO skins treated with NAC (or vehicle) for 48 h, of three mice/genotype; *P<0.05, **P<0.005. (e) Histograms represent mean±S.D. of TUNEL+ cells/area (103 μm2) of at least 30 hair follicles of 10 mice/genotype treated as in c; ***P<0.0005.
We next analyzed in CT and E-RiKO epidermis the expression of a sample of DEGs between CT and E-RiKO keratinocytes emerged from RNA-seq analysis. As shown in Figure 7b, the levels of Gadd45α, -β and -γ, Glul and Il1-α genes, involved in DNA repair, glutamine metabolism and inflammation, upregulated in E-RiKO cells (Supplementary Table S1), were also higher in the mutant epidermis, whereas the levels of Acsl1, Faah and Fabp4, involved in lipid metabolism, were decreased.
To determine whether NAC treatment could sensitize E-RiKO hair follicle keratinocytes to X-ray-induced death in vivo, we established a NAC treatment regimen restoring ROS levels in mutant mice to levels close to CTs (Figure 7d). As shown in Figures 7c–e, NAC-treated E-RiKO mice exhibited a statistically significant increase in the number of TUNEL+ cells in the hair follicle matrix upon irradiation, suggesting that restoring normal ROS levels in E-RiKO mice sensitizes hair follicle keratinocytes to X-ray-induced cell death in vivo. Moreover, NAC slightly increased the number of TUNEL+ CT cells (Figure 7e and data not shown). Thus, many aspects of the metabolic, molecular and stress-response phenotypes described in primary keratinocytes derived from E-RiKO mice are recapitulated in their intact skin epithelia in vivo.
Discussion
Our study reveals for the first time that mTORC2 couples metabolic changes with stress adaptation in mammalian cells. The resiliency of rictor-deficient keratinocytes to starvation, epirubicin and X-ray in vitro and their radioresistance in vivo was somewhat surprising, considering the pro-survival role of mTORC2 in mammals.15 However, mTORC2 deficiency does not render keratinocytes resistant to cell death per se but likely enhances their ability to cope with oxidative stress as these cellular stressors share ROS production among their death effector mechanisms.27 Because in control keratinocytes, Akt Ser473 phosphorylation decreases earlier upon X-ray (but not cisplatin) treatment, attenuation of mTORC2 activity may be intrinsic to the keratinocyte response to specific stressors.
Our findings in mammalian cells have intriguing analogies in Drosophila, where rictor deficiency promotes resistance and tolerance to pathogens infection under low glucose,36 suggesting a conserved role of mTORC2 in integrating metabolic cues and stress adaptation. As the mTORC2/Akt axis positively regulates glucose uptake and glycolysis in several tissues,22, 37 the stress-resistant phenotype of E-RiKO cells may result from their adaptation to impaired glucose consumption and rewiring towards glutaminolysis. The increased glutamine consumption of E-RiKO keratinocytes is associated with increased activity of glutaminolytic enzymes without changes in their protein or mRNA levels suggesting that rictor deficiency may affects these enzymes via post-translational modifications or by changing the concentration of allosteric modulations, as it occurs on mTORC2 disruption.38 In addition, rictor loss may impinge on Foxo transcription factors in linking metabolic changes with stress resistance. Foxo3 regulates glutamine metabolism via Glul expression;39 Glul is overexpressed in E-RiKO cells that also have reduced Foxos phosphorylation, and among stress protective genes overexpressed in E-RiKO cells are the Foxo targets Gadd45 isoforms.40
Mitochondrial SOD enzymatic activity, having a pivotal role in radioprotection,41 is elevated in E-RiKO cells, and decreases upon NAC treatment. Active SOD2 is stabilized by oxidative cross-linking, favoring radioresistance of cells exposed to conditioning doses of oxidative stress.42 Enhanced SOD2 expression accounts for radioresistance of rapamycin-treated oral keratinocytes,41 and SOD2 activity can be regulated post-translationally in response to ROS levels.42 Therefore, changes in SOD2 activity likely favor stress adaptation in rictor-deficient keratinocytes, albeit the underlying mechanisms remain undefined.
The increased ROS levels of E-RiKO keratinocytes, instead of being harmful, emerged as key determinants of their stress-resistant phenotype. Therefore, the behavior of rictor-deficient keratinocytes fits well with the concept of mitohormesis, implicating mTORC2 in radioresistance, and possibly, skin aging. Hormetic behaviors occur in several cell lineages including keratinocytes, and mild level of X-ray render cells more resistant to a subsequent damage.43
Our findings overall suggest that the switch of keratinocyte catabolism towards glutaminolysis provides fuel for mitochondrial respiration and ROS production, which, directly or indirectly, induces expression of stress resistance genes.
Rictor deficiency was associated with decreased cell division18, 44 and the attenuated proliferation of E-RiKO keratinocytes likely underlies the epidermal hypoplasia of mutant mice, albeit we cannot rule out subtle defects in differentiation or cytoskeleton dynamics. Adult E-RiKO mice epidermis does not display hypoplasia or impaired proliferation unless challenged with a mitogen like TPA; in growth factor-rich conditions, E-RiKO keratinocytes also exhibit reduced division rates, suggesting that mTORC2 loss affects proliferation upon robust mitogenic inputs. The attenuated hyperplastic response to one single TPA dose reported here differs from the normal hyperplastic response to multiple TPA doses described in mice subjected to epidermal-specific, inducible rictor ablation during adulthood.45 Metabolic alterations coupled with enhanced stress tolerance likely contribute to the delayed senescence and lifespan extension of rictor-deficient keratinocytes, in keeping with similar reports on fibroblasts.46 As hyperactive DNA replication forks promote senescence, the spontaneous immortalization of E-RiKO keratinocytes may result from senescence bypass favored by metabolic reprogramming, mitohormesis and reduced proliferation rates. As sustained exposure of E-RiKO keratinocytes to NAC enhances BrdU uptake and favors senescence in the absence of exogenous stressors, ROS elevations may contribute to the proliferative and senescence phenotypes of mutant cells.
Mitohormesis may represent a point of convergence of mTORC1 and mTORC2 activities across species. In fact, mTORC1 inhibition extends yeast chronological lifespan;11 the mTOR inhibitor rapamycin extends lifespan in C. elegans and mice47, 48 and prolonged rapamycin exposure also inhibits mTORC2;48, 49 rictor deficiency in yeast was linked to increased ROS levels,50 and in C. elegans, rictor mutations extend lifespan in a nutrient-dependent manner.51
The choice between cell survival and death upon mTORC2 disruption may be highly context-dependent. Rictor/mTORC2 deficiency sensitizes fibroblasts to UVB-induced apoptosis,29 but the mechanisms regulating DNA repair and apoptosis differ between keratinocytes and fibroblasts,52 possibly accounting for this apparent discrepancy. In addition, in breast cancer cells, mTORC2 loss favors cell cycle progression and apoptosis by lowering Chk1 activity and increasing vulnerability to DNA damage;29 in E-RiKO cells Chk1 attenuation coincides with lower DNA damage in response to X-ray, but not cisplatin. mTOR inhibition can inhibit or enhance chemotherapy-induced cancer cell death in oncological settings;53 if mTOR inhibition enhances stress tolerance via mitohormesis, this could partly explain the limited efficacy of mTOR inhibitors in this context.54 mTORC2 disruption inhibits tumor formation/progression in many tissues including the epidermis,45 but caution should be taken in therapeutic settings as mTOR inhibition may also promote chemo- or radioresistance. Our work suggests, however, that combination of mTOR inhibitors with antioxidants may help sensitizing tumors that would be otherwise resistant to chemo/radio-therapy.
Materials and methods
Mice generation and genotyping
Generation of rictor conditional knockout mice in the epidermis (E-RiKO) was obtained by crossing Rictor flox/flox mice (CT) described in ref. 17 with K14-Cre transgenic mice.23 The mice were studied on a C57BL/6J background. Genetic screening was performed by PCR using DNA extracted from tail biopsies.
The primer used for genotyping were as follows:
-
Rictor flox Forward, 5′-ACTGAATATGTTCATGGTTGTG-3′
-
Rictor flox Reverse, 5′-GACACTGGATTACAGTGGCTTG-3′
-
K14-cre Forward, 5′-AGGGATCTGATCGGGAGTTG-3′
-
K14-cre Reverse, 5′-CTTGCGAACCTCATCACTCG-3′
The mice were maintained under temperature- and humidity-controlled conditions and were given food and water ad libitum. The procedures were conducted in conformity with national and international laws and policies as approved by the University of Turin Ethical Committee.
Cell cultures and chemicals
MPKs were isolated from pools of 3-day-old (P3) mice and cultured in low calcium medium (LCM, 50 μM CaCl2 supplemented with 4% Chelex-treated bovine serum and EGF). The cultures were ~95% pure and contained traces of melanocytes and Langerhans’ cells. MPKs cultures were obtained from at least five mice/genotype and, unless otherwise specified, all the experiments were repeated on at least three independent cultures.
Note that MPKs cultured in LCM keep proliferating even at confluency because they are preserved from contact inhibition, and dividing cells replace the differentiated ones that lift from the adherent cell monolayer.
Starvation was performed culturing MPKs in serum and EGF-free LCM, except for kinetic analysis of BrdU incorporation, in which cells were starved in 0.1% serum.
The cells fed with LCM (or starved) were treated with the following chemicals: epirubicin, cisplatin and NAC (Sigma-Aldrich, St. Louis, MO, USA), Trolox (Santa Cruz Biotechnology, Dallas, TX, USA) and BPTES (Selleckchem, Houston, TX, USA).
NAC treatments were performed by adding the compound into fresh LCM every 24 h. To evaluate proliferation, MPKs were incubated with 5-bromo-2″-deoxy-uridine (BrdU, Invitrogen, Waltham, MA, USA) 3 h before the end of the experiment (10 μg/ml) and then fixed in PFA 4%.
Colony-forming efficiency (CFE) assays were performed as described,55 with minor modifications. After X-ray exposure (10 min), the cells were detached, and 104 cells were plated on lethally irradiated feeder layer of 3T3/J2 cells. After 8 days, colonies were fixed, stained with Rhodamine-B (Sigma-Aldrich) and scored under a dissecting microscope. Total colonies were calculated as a percentage of total plated cells (number of colonies × 100/number of cells plated).
Western blotting and protein analysis
MPKs or epidermal tissues (P3), separated from dermis by thermal shock at 65 °C for 1 min, were lysed in boiling 2%SDS, 50 mM Tris/HCl (pH 7.4) lysis buffer supplemented with 1 mM PMSF, 1 mM Na3VO4, 10 mM NaF.41 Epidermal tissues were pulverized in liquid nitrogen before lysis. Protein concentration was measured using the Bradford assay (Bio-Rad, Hercules, CA, USA). The samples were fractionated on SDS/PAGE and transferred on to PVDF membrane (Merk Millipore, Billerica, MA, USA). The membranes were blocked in 5% non-fat dry milk (Santa Cruz Biotechnology) in Tris-buffer saline, 0.1% Tween20 and incubated with the indicated antibodies following the manufacturer’s instructions.
For immunoblotting, the following antibodies were used: rictor, raptor, pSer473Akt, pThr308Akt, panAkt, p-GSK3 α/β (Ser 21/9), p-FOXO1/3 (Thr32/Thr24), pFoxO1 (Ser256, cross-reactive with Ser193 of FoxO4), pFoxO3 (Ser253), FoxO1, FoxO3, p-TSC2 (Thr1462), total TSC2, mTOR, p-p70S6K (Thr389), total p70S6K, p-4EBP1, total 4EBP1, pPRAS40 (Thr246), p-Erk1/2 (Thr37/46), total Erk1/2, Cleaved Caspase-3, pChk1 (Ser317), total Chk1, pChk2 (Thr68), total Chk2 (Cell Signaling Technology, Danvers, MA, USA); PKCα, Hsp90, p16, p19, p-Ser19 p53, p53 (Santa Cruz Biotechnology); SOD1, SOD2, catalase (Abcam, Cambridge, UK) GAPDH (Merck-Millipore); Vinculin, Tubulin (Sigma-Aldrich); K5 (Covance, Princeton, NJ, USA) and horseradish peroxidase-conjugated secondary antibodies (Sigma-Aldrich). Immunoblots were developed by chemiluminescence with ECL (GE Healthcare, Dharmacon, Lafayette, CO, USA), acquired with the molecular imager ChemiDoc XRS, and quantified by densitometric analysis using the Image-lab software (Bio-Rad). All comparative images of immunoblots were obtained by exposure of the same membranes. Original immunoblots are shown in Supplementary Figures S6 and S7.
Treatments in vivo
For the acute response to 12-O-tetradecanoylphorbol-13-acetate (TPA), dorsal skin of 7-week-old mice of both genotypes were shaved and treated with a single dose of TPA (0.1 mM, Sigma-Aldrich) or acetone vehicle alone.56 The treated mice were killed after 48 h for histopathological analysis.
For BrdU incorporation analysis in response to TPA, the mice were i.p. injected with 50 mg/kg of BrdU in sterile PBS, 1 h before the termination of experiments.
The mice (P3) were X-ray irradiated (full body) with 4 or 8 Gy doses and killed for skin analysis 24 h later.
For NAC treatment, CT and E-RiKO mice (P2) were i.p. injected with NAC (Sigma) in PBS sterile solution (100 mg/kg) or PBS alone daily for 3 days and then irradiated full body (8 Gy). The mice were killed and their skins were excised 24 h after X-ray exposure.
Immunofluorescence and immunohistochemistry
The skins were fixed in 4% buffered formalin for 24 h and embedded in paraffin. Seven-micrometer-thick skin sections were analyzed as follows: hematoxylin and eosin (H&E) staining; IHC was performed for BrdU (DAKO, Carpinteria, CA, USA) and PCNA (Santa Cruz Biotechnology); IF was performed for p63 (Santa Cruz Biotechnology), Loricrin (Covance), TUNEL (Roche, Basel, Switzerland) following the manufacturer’s instructions. The IF samples were counterstained with DAPI or LaminA (Santa Cruz Biotechnology) and mounted in Prolongue reagent (Life Technologies, Carlsbad, CA, USA).
Flow cytometry
The dead and viable cells were estimated based on AnnexinV/DAPI staining followed by flow cytometry. For this purpose, both adherent and spontaneously detached keratinocytes in each condition were incubated with AnnV-FITC (BD Biosciences) for 30 min in the dark at room temperature and DAPI (Sigma) was added right before the measurement (1 μg/ml). The cells were distinguished in live (double negative) and dead: early apoptotic (AnnV+DAPI−), medium/late apoptotic (AnnV+DAPI+) or necrotic (AnnV−DAPI+). Flow cytometric data were acquired using a FACSVerse (Becton Dickinson) and processed with FACSuite software. At least 20 000 events were analyzed for each sample.
The dead cells were calculated by setting to 100% the mean of treated (starvation, epirubicin, X-ray, cisplatin, Nac/epirubicin, BPTES/epirubicin) CT dead cells, obtained by subtracting untreated CT dead cells. The variation of dead cell fraction for each stressor was calculated taking the ratio of treated E-RiKO dead cells (corrected by subtracting untreated E-RiKO dead cells) to treated CT dead cells. Data are representative of at least three independent experiments.
Senescence-associated β-galactosidase assay
MPKs were plated in triplicate, cultured for 25 days and stained for β-galactosidase activity using the Senescence Detection Kit (Cell Signaling and Technology) following the manufacturer’s instructions.
X-ray irradiation
Irradiation was done using a Gilardoni RADGIL irradiator (Stationary anode X-ray tube, 200 kV) at 0.65 Gy/min. MPKs were irradiated at subconfluency in LCM.
RNA-Seq
Total RNA was extracted using TRIZOL reagent (Invitrogen) and checked for its integrity by using the DNF-471 Standard Sensitivity RNA Analysis Kit on Fragment Analyzer instrument (Advanced Analytical Technology, Ankeny, IA, USA). RNA-seq libraries were prepared from total RNA using TruSeq RNA Sample Preparation v2 (Illumina, San Diego, CA, USA) according to the manufacturer's protocol and were sequenced on Illumina NextSeq 500 platform (Illumina). Sequencing reads were trimmed out of the low-quality bases with Fastx Toolkit and were mapped on hg19 genome assembly by using TopHat v2.0.6 (Johns Hopkins University, Baltimore, MD, USA) and mRNAs quantification were performed using Cuffdiff v2.0.2 (University of Maryland, College Park, MD, USA). For further analysis, genes with RPKM<1 in all the samples were filtered out. Gene Ontology was analyzed by using GO web software.
Biochemical analysis of cell metabolism
Mitochondria extraction
To isolate mitochondrial fractions, the cells or pulverized skins were washed twice in ice-cold PBS, lysed in 0.5 ml mitochondria lysis buffer (50 mmol/l Tris, 100 mmol/L KCl, 5 mmol/l MgCl2, 1.8 mmol/l ATP, 1 mmol/l EDTA, pH 7.2), supplemented with protease inhibitor cocktail III (Calbiochem, La Jolla, CA, USA), 1 mmol/l PMSF and 250 mmol/l NaF. The samples were clarified by centrifuging at 650 g for 3 min at +4 °C: the supernatant was collected and centrifuged at 13 000 g for 5 min at +4 °C; the pellet – containing mitochondria – was washed once with lysis buffer and resuspended in 0.25 ml mitochondria resuspension buffer (250 mmol/l sucrose, 15 mmol/l K2HPO4, 2 mmol/l MgCl2, 0.5 mmol/l EDTA). A 50 μl aliquot was sonicated and used for the measurement of protein content or western blotting. To confirm the presence of mitochondrial proteins in the extracts, 10 μg of each sonicated sample were subjected to SDS-PAGE and probed with an anti-porin antibody (Abcam, Cambridge, UK; data not shown).
ROS measurement
ROS amount in whole cells or in mitochondria extracts was measured by labeling samples with the ROS-sensitive fluorescent probe 5-(and-6)-chloromethyl-2',7'-dichlorodihydro-fluorescein diacetate-acetoxymethyl ester (DCFDA-AM). The results were expressed as nmol/mg cell or mitochondrial proteins.
Aldose reductase and NADPH oxidase activity
The activities of aldose reductase and NADPH oxidase were measured by a spectrophotometric assay and by a chemiluminscence-based assay, respectively.57 The results were expressed as nmoles NADP+/min/mg cell proteins for aldose reductase, relative luminescence unit (RLU)/mg cell proteins for NADPH oxidase.
Mitochondrial respiratory chain
To measure the electron flux from complex I to complex III, taken as index of the mitochondrial respiratory activity, 50 μg of non-sonicated mitochondrial samples, isolated as previously reported, were resuspended in 0.2 ml buffer A (5 mmol/l KH2PO4, 5 mmol/l MgCl2, 5% w/v bovine serum albumin) and transferred into a quartz spectrophotometer cuvette. Then 0.1 ml buffer B (25% w/v saponin, 50 mmol/l KH2PO4, 5 mmol/l MgCl2, 5% w/v bovine serum albumin, 0.12 mmol/l cytochrome c-oxidized form, 0.2 mmol/l NaN3) was added for 5 min at room temperature. The reaction was started with 0.15 mmol/l NADH and was followed for 5 min, reading the absorbance at 550 nm by a Packard microplate reader EL340 (Bio-Tek Instruments, Winooski, VT, USA). The results were expressed as nanomoles of cytochrome c reduced/min/mg mitochondrial protein.
ATP levels measurement
The amount of ATP in mitochondrial extracts was measured with the ATP Bioluminescent Assay Kit (Sigma-Aldrich). The results were expressed as nmol/mg mitochondrial proteins.
Triglycerides levels
The triglyceride amount was measured using the Triglyceride Quantification Kit (Abcam), following the manufacturer’s instruction. The results were expressed in nmol/mg cell or tissue proteins, according to the calibration curve previously set.
Fatty acid β-oxidation
The rate of fatty acid β-oxidation was measured by radiolabeling cells or pulverized skins with 2 μCi [1-14C]palmitic acid (3.3 mCi/mmol; PerkinElmer, Waltham, MA, USA) and quantifying the amount of 14C-acid soluble metabolites (ASM) by liquid scintillation.57 The results were expressed as picomoles of 14C-ASM/h/mg cell proteins.
Glucose uptake and glycolytic metabolism
The uptake of glucose was measured as described earlier58 and expressed as picomoles of 2-deoxy-D-[3H]-glucose/mg cell proteins. HK activity was measured by using the Hexokinase Colorimetric Assay Kit (Sigma-Aldrich). The results were expressed as nanomoles of NADH/min/mg cell proteins. PFK1 assay was performed according to ref. 59. The results were expressed as nanomoles of NAD+/min/mg cell proteins.
Glutamine catabolism
Glutamine catabolism was measured as reported.57 The cells or pulverized skins were washed with PBS, detached by gentle scraping, centrifuged at 13 000 × g for 5 min at 4 °C, resuspended in 250 μl of buffer A (150 mmol/l KH2PO4, 63 mmol/l Tris/HCl, 0.25 mmol/l EDTA; pH 8.6) and sonicated. The intracellular protein content was measured using the BCA kit (Sigma Chemical Co.). A volume of 100 μl of the whole cell lysates was incubated for 30 min at 37 °C in a quartz cuvette, in the presence of 50 μl of 20 mmol/l l-glutamine and 850 μl of buffer B (80 mmol/l Tris/HCl, 20 mmol/l NAD+, 20 mmol/l ADP, 3% v/v H2O2; pH 9.4). The absorbance of NADH was monitored at 340 nm using a Lambda 3 spectrophotometer (PerkinElmer). The kinetics was linear throughout the assay. The results were expressed as micromoles of NADH/min/mg cell proteins, and were considered as an index of the activity of glutaminase plus l-glutamic dehydrogenase. In a second series of samples, 20 μl of the glutaminase inhibitor bis-2-(5–phenylacetamido–1,3,4–thiadiazol–2–yl)ethyl sulfide BTPES (30 μmol/l) was added after 15 min. This concentration was chosen as it produced 100% inhibition of glutaminase activity in our system (not shown). The absorbance of NADH was monitored for 15 min as described previously. The results, considered as an index of the activity of l-glutamic dehydrogenase, were expressed as micromoles of NADH/min/mg cell proteins. Glutaminase activity was obtained by subtracting the rate of the second assay from the rate of the first one.
Tricarboxylic acid (TCA) cycle
The cells were washed with PBS, detached with trypsin/EDTA (0.05/0.02% v/v) and resuspended in 1 ml Hepes buffer (145 mmol/l NaCl, 5 mmol/l KCl, 1 mmol/l MgSO4, 10 mmol/l Hepes, 10 mmol/l glucose, 1 mmol/l CaCl2, pH 7.4) containing 2 μCi of l-[14C]-glutamine (PerkinElmer) or l-[14C]-glutamate (PerkinElmer). The cell suspensions were incubated for 1 h in a closed experimental system to trap the 14CO2 developed from l-[14C]-glutamine or l-[14C]-glutamate and the reaction was stopped by injecting 0.5 ml 0.8 N HClO4. The results were expressed as nanomoles of CO2/h/mg cell proteins. When indicated, 30 μmol/l of the glutaminase inhibitor BPTES were added to the cell suspension, to achieve a 100% inhibition of glutaminase activity.
Glutamine consumption
The cells were labeled with 1 μCi [14C]-l-glutamine (PerkinElmer) for 30 min, washed five times with ice-cold PBS, detached with trypsin/EDTA, rinsed with 0.5 ml ice-cold PBS and sonicated. A 50 μl aliquot was used to quantify intracellular proteins. [14C]-l-glutamate and [14C]-l-glutamine present within cell lysates were separated by ion exchange chromatography in a 2 ml column. The radioactivity of the eluate containing [14C]-l-glutamate and [14C]-l-glutamine was counted by liquid scintillation and expressed as μmol/mg cellular proteins. The ratio between [14C]-l-glutamate/[14C]-l-glutamine was considered an index of glutamine consumption.
Real-time PCR
Total RNA was extracted from the epidermis of mice (P3) using Triazol reagent (Invitrogen) according to the manufacturer’s instructions. Total RNA was reverse-transcribed with a high-capacity cDNA reverse transcription kit (Applied BioSystems, Foster city, CA, USA) according to the manufacturer's instruction and amplified with specific primers. Taqman PCR reactions were performed using the Universal Probe Library system (Roche Italia, Monza, Italy) on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). The 18 S rRNA pre-developed TaqMan assay (Applied Biosystems) was used as an internal control. Specific primers and UPL probes used are listed in Supplementary Informations.
Microscopy and image analysis
Immunofluorescence analysis was performed on Leica TSCII SP5 confocal microscope (Leica, Wetzlar, Germany) controlled by LAS-AF Software (Leica). Multitrack analysis was used for image acquisition. Histological sections were imaged on Olympus BH-2 RFCA microscope equipped with Leica DFC320 camera (Leica). Camera was controlled by Leica Application Suite version 2.8.1 software (Leica). Phase contrast imaging was performed on Zeiss Axio Observer microscope (Carl Zeiss, Oberkochen, Germany). Quantitative analysis was performed using ImageJ software (National Institute of Health, Bethesda, MD, USA).
Statistical analysis
The data obtained from densitometric analysis of immunoblots, FACS, IF, CFE and IHC were plotted as mean±S.D. The results were assessed for statistical significance by a standard two-tailed Student’s t-test as indicated; P-values *P<0.05, **P<0.005, ***P<0.0005. For epidermal thickness, measurements obtained from H&E were analyzed with linear regression using Generalized Estimating Equations (GEE)60 to take into account the correlation between measurements obtained from the same animal. Data obtained from growth assays were analyzed by using the CompareGrowthCurves function included in the R package ‘statmod’: https://cran.r-project.org/web/packages/statmod/index.html
References
Blanpain C, Fuchs E . Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 2009; 10: 207–217.
Brandhorst S, Choi IY, Wei M, Cheng CW, Sedrakyan S, Navarrete G et al. A periodic diet that mimics fasting promotes multi-system regeneration, enhanced cognitive performance, and healthspan. Cell Metab 2015; 22: 86–99.
Ristow M, Schmeisser K . Mitohormesis: promoting health and lifespan by increased levels of reactive oxygen species (ROS). Dose Response 2014; 12: 288–341.
Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M . Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab 2007; 6: 280–293.
Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D et al. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab 2012; 15: 451–465.
Sena LA, Chandel NS . Physiological roles of mitochondrial reactive oxygen species. Mol Cell 2012; 48: 158–167.
Strozyk E, Kulms D . The role of AKT/mTOR pathway in stress response to UV-irradiation: implication in skin carcinogenesis by regulation of apoptosis, autophagy and senescence. Int J Mol Sci 2013; 14: 15260–15285.
Soeur J, Eilstein J, Lereaux G, Jones C, Marrot L . Skin resistance to oxidative stress induced by resveratrol: from Nrf2 activation to GSH biosynthesis. Free Radic Biol Med 2015; 78: 213–223.
Angel P, Szabowski A, Schorpp-Kistner M . Function and regulation of AP-1 subunits in skin physiology and pathology. Oncogene 2001; 20: 2413–2423.
Zoncu R, Efeyan A, Sabatini DM . mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011; 12: 21–35.
Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS . Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab 2011; 13: 668–678.
Wullschleger S, Loewith R, Hall MN . TOR signaling in growth and metabolism. Cell 2006; 124: 471–484.
Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell 2006; 11: 859–871.
Dann SG, Selvaraj A, Thomas G . mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med 2007; 13: 252–259.
Sarbassov DD, Guertin DA, Ali SM, Sabatini DM . Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005; 307: 1098–1101.
Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 2004; 6: 1122–1128.
Shiota C, Woo JT, Lindner J, Shelton KD, Magnuson MA . Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev Cell 2006; 11: 583–589.
Wang S, Amato KR, Song W, Youngblood V, Lee K, Boothby M et al. Regulation of endothelial cell proliferation and vascular assembly through distinct mTORC2 signaling pathways. Mol Cell Biol 2015; 35: 1299–1313.
Thomanetz V, Angliker N, Cloetta D, Lustenberger RM, Schweighauser M, Oliveri F et al. Ablation of the mTORC2 component rictor in brain or Purkinje cells affects size and neuron morphology. J Cell Biol 2013; 201: 293–308.
Kumar A, Harris TE, Keller SR, Choi KM, Magnuson MA, Lawrence JC Jr . Muscle-specific deletion of rictor impairs insulin-stimulated glucose transport and enhances Basal glycogen synthase activity. Mol Cell Biol 2008; 28: 61–70.
Yuan M, Pino E, Wu L, Kacergis M, Soukas AA . Identification of Akt-independent regulation of hepatic lipogenesis by mammalian target of rapamycin (mTOR) complex 2. J Biol Chem 2012; 287: 29579–29588.
Kumar A, Lawrence JC Jr, Jung DY, Ko HJ, Keller SR, Kim JK et al. Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes 2010; 59: 1397–1406.
Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W . beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell 2001; 105: 533–545.
Brown J, Wang H, Suttles J, Graves DT, Martin M . Mammalian target of rapamycin complex 2 (mTORC2) negatively regulates Toll-like receptor 4-mediated inflammatory response via FoxO1. J Biol Chem 2011; 286: 44295–44305.
Li J, Foitzik K, Calautti E, Baden H, Doetschman T, Dotto GP . TGF-beta3, but not TGF-β1, protects keratinocytes against 12-O-tetradecanoylphorbol-13-acetate-induced cell death in vitro and in vivo. J Biol Chem 1999; 274: 4213–4219.
Orecchia V, Regis G, Tassone B, Valenti C, Avalle L, Saoncella S et al. Constitutive STAT3 activation in epidermal keratinocytes enhances cell clonogenicity and favours spontaneous immortalization by opposing differentiation and senescence checkpoints. Exp Dermatol 2015; 24: 29–34.
Simunek T, Sterba M, Popelova O, Adamcova M, Hrdina R, Gersl V . Anthracycline-induced cardiotoxicity: overview of studies examining the roles of oxidative stress and free cellular iron. Pharmacol Rep 2009; 61: 154–171.
Petit-Frere C, Capulas E, Lyon DA, Norbury CJ, Lowe JE, Clingen PH et al. Apoptosis and cytokine release induced by ionizing or ultraviolet B radiation in primary and immortalized human keratinocytes. Carcinogenesis 2000; 21: 1087–1095.
Carr TD, DiGiovanni J, Lynch CJ, Shantz LM . Inhibition of mTOR suppresses UVB-induced keratinocyte proliferation and survival. Cancer Prev Res (Phila) 2012; 5: 1394–1404.
Sotiropoulou PA, Candi A, Mascre G, De Clercq S, Youssef KK, Lapouge G et al. Bcl-2 and accelerated DNA repair mediates resistance of hair follicle bulge stem cells to DNA-damage-induced cell death. Nat Cell Biol 2010; 12: 572–582.
Song S, Lambert PF . Different responses of epidermal and hair follicular cells to radiation correlate with distinct patterns of p53 and p21 induction. Am J Pathol 1999; 155: 1121–1127.
Siddik ZH . Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 2003; 22: 7265–7279.
Jiang F, Zhang Y, Dusting GJ . NADPH oxidase-mediated redox signaling: roles in cellular stress response, stress tolerance, and tissue repair. Pharmacol Rev 2011; 63: 218–242.
Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 2013; 496: 101–105.
Yamada T, Egashira N, Bando A, Nishime Y, Tonogai Y, Imuta M et al. Activation of p38 MAPK by oxidative stress underlying epirubicin-induced vascular endothelial cell injury. Free Radic Biol Med 2012; 52: 1285–1293.
Allen VW, O'Connor RM, Ulgherait M, Zhou CG, Stone EF, Hill VM et al. Period-regulated feeding behavior and TOR signaling modulate survival of infection. Curr Biol 2016; 26: 184–194.
Albert V, Svensson K, Shimobayashi M, Colombi M, Munoz S, Jimenez V et al. mTORC2 sustains thermogenesis via Akt-induced glucose uptake and glycolysis in brown adipose tissue. EMBO Mol Med 2016; 8: 232–246.
Moloughney JG, Kim PK, Vega-Cotto NM, Wu CC, Zhang S, Adlam M et al. mTORC2 responds to glutamine catabolite levels to modulate the hexosamine biosynthesis enzyme GFAT1. Mol Cell 2016; 63: 811–826.
van der Vos KE, Eliasson P, Proikas-Cezanne T, Vervoort SJ, van Boxtel R, Putker M et al. Modulation of glutamine metabolism by the PI(3)K-PKB-FOXO network regulates autophagy. Nat Cell Biol 2012; 14: 829–837.
Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ Jr, DiStefano PS et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science 2002; 296: 530–534.
Iglesias-Bartolome R, Patel V, Cotrim A, Leelahavanichkul K, Molinolo AA, Mitchell JB et al. mTOR inhibition prevents epithelial stem cell senescence and protects from radiation-induced mucositis. Cell Stem Cell 2012; 11: 401–414.
Belikova NA, Glumac A, Rafikov R, Jiang J, Greenberger JS, Kagan VE et al. Radioprotection by short-term oxidative preconditioning: role of manganese superoxide dismutase. FEBS Lett 2009; 583: 3437–3442.
Berge U, Kristensen P, Rattan SI . Hormetic modulation of differentiation of normal human epidermal keratinocytes undergoing replicative senescence in vitro. Exp Gerontol 2008; 43: 658–662.
Gu Y, Lindner J, Kumar A, Yuan W, Magnuson MA . Rictor/mTORC2 is essential for maintaining a balance between beta-cell proliferation and cell size. Diabetes 2011; 60: 827–837.
Carr TD, Feehan RP, Hall MN, Ruegg MA, Shantz LM . Conditional disruption of rictor demonstrates a direct requirement for mTORC2 in skin tumor development and continued growth of established tumors. Carcinogenesis 2015; 36: 487–497.
Zelenka J, Dvorak A, Alan L . L-lactate protects skin fibroblasts against aging-associated mitochondrial dysfunction via mitohormesis. Oxid Med Cell Longev 2015; 2015: 351698.
Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009; 460: 392–395.
Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E et al. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab 2012; 15: 713–724.
Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 2006; 22: 159–168.
Niles BJ, Joslin AC, Fresques T, Powers T . TOR complex 2-Ypk1 signaling maintains sphingolipid homeostasis by sensing and regulating ROS accumulation. Cell Rep 2014; 6: 541–552.
Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G . Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev 2009; 23: 496–511.
D'Errico M, Teson M, Calcagnile A, Nardo T, De Luca N, Lazzari C et al. Differential role of transcription-coupled repair in UVB-induced response of human fibroblasts and keratinocytes. Cancer Res 2005; 65: 432–438.
Selvarajah J, Nathawat K, Moumen A, Ashcroft M, Carroll VA . Chemotherapy-mediated p53-dependent DNA damage response in clear cell renal cell carcinoma: role of the mTORC1/2 and hypoxia-inducible factor pathways. Cell Death Dis 2013; 4: e865.
Lee JS, Vo TT, Fruman DA . Targeting mTOR for the treatment of B cell malignancies. Br J Clin Pharmacol 2016; 82: 1213–1228.
Saoncella S, Tassone B, Deklic E, Avolio F, Jon C, Tornillo G et al. Nuclear Akt2 opposes limbal keratinocyte stem cell self-renewal by repressing a FOXO-mTORC1 signaling pathway. Stem Cells 2014; 32: 754–769.
Li J, Wei Y, Wagner TE . In vitro endothelial differentiation of long-term cultured murine embryonic yolk sac cells induced by matrigel. Stem Cells 1999; 17: 72–81.
Capello M, Ferri-Borgogno S, Riganti C, Chattaragada MS, Principe M, Roux C et al. Targeting the Warburg effect in cancer cells through ENO1 knockdown rescues oxidative phosphorylation and induces growth arrest. Oncotarget 2015; 7: 5598–5612.
Bergandi L, Silvagno F, Russo I, Riganti C, Anfossi G, Aldieri E et al. Insulin stimulates glucose transport via nitric oxide/cyclic GMP pathway in human vascular smooth muscle cells. Arterioscler Thromb Vasc Biol 2003; 23: 2215–2221.
Sharma B . Kinetic characterisation of phosphofructokinase purified from Setaria cervi: a bovine filarial parasite. Enzyme Res 2011; 2011: 939472.
Hanley JA, Negassa A, Edwardes MD, Forrester JE . Statistical analysis of correlated data using generalized estimating equations: an orientation. Am J Epidemiol 2003; 157: 364–375.
Acknowledgements
We apologize to the many colleagues whose primary work could not be cited directly due to space constraints. We are thankful to Dr J Huelsken for making available K14-Cre transgenic mice. We especially thank PP Pandolfi for constant feedback and fruitful discussions and Mara Brancaccio, Valeria Poli, Emilio Hirsch, Alessandra Ghigo for critical reading of the manuscript. This work was supported by Telethon Foundation, Italy (TCP 06001 to EC) and by the Italian Association for Cancer Research (AIRC; grant IG 15232 to CR). We especially thank Professor F Altruda for precious suggestions and support (Prometeo grant ALTF_RIC_ACT_15_01).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by E Gottlieb
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Tassone, B., Saoncella, S., Neri, F. et al. Rictor/mTORC2 deficiency enhances keratinocyte stress tolerance via mitohormesis. Cell Death Differ 24, 731–746 (2017). https://doi.org/10.1038/cdd.2017.8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2017.8
This article is cited by
-
Mitochondrial ROS drive resistance to chemotherapy and immune-killing in hypoxic non-small cell lung cancer
Journal of Experimental & Clinical Cancer Research (2022)
-
Multifunctional thiosemicarbazones targeting sigma receptors: in vitro and in vivo antitumor activities in pancreatic cancer models
Cellular Oncology (2021)
-
Sublethal exposure of small few-layer graphene promotes metabolic alterations in human skin cells
Scientific Reports (2020)