Abstract
Sustained activation of nuclear factor-κB (NF-κB) in cancer cells has been shown to promote inflammation, expansion of cancer stem cell (CSC) population, and tumor development. In contrast, recent studies reveal that CSCs exhibit increased inflammation due to constitutive NF-κB activation; however, the underlying molecular mechanism remains unclear. In the present study, the analysis of microarray data revealed upregulation of NF-κB-regulated pro-inflammatory genes and downregulation of copper metabolism MURR1 domain-containing 1 (COMMD1) during the enrichment for stemness in SAS head and neck squamous-cell carcinoma (HNSCC) cells. The 3′-UTR of COMMD1 mRNA contains microRNA (miR)-205 target site. Parallel studies with HNSCC and NSCLC cells indicated that miR-205 is upregulated upon NF-κB activation and suppresses COMMD1 expression in stemness-enriched cancer cells. COMMD1 negatively regulates the inflammatory responses induced by TLR agonists, IL-1β, and TNF-α by targeting RelA for degradation. The shRNA-mediated downregulation of COMMD1 in cancer cells enhanced inflammatory response, generating favorable conditions for macrophage recruitment. In addition, genes associated with stemness were also upregulated in these cells, which exhibited increased potential for anchorage-independent growth. Furthermore, COMMD1 downregulation promoted in vivo tumorigenesis and tumor growth, and tumors derived from COMMD1-knockdown cells displayed elevated level of NF-κB activation, increased expression of inflammatory- and stemness-associated genes, and contain expanded population of tumor-associated leukocytes and stemness-enriched cancer cells. These results suggest that COMMD1 downregulation by miR-205 promotes tumor development by modulating a positive feedback loop that amplifies inflammatory- and stemness-associated properties of cancer cells.
Similar content being viewed by others
Main
Accumulating evidence supports the association between inflammation and tumorigenesis. Persistent inflammatory response induced by extrinsic factors such as exposure to environmental agents, infection with a pathogen, genetic disease, or metabolic disorder increases the predisposition of patients with subclinical conditions to cancer. Inflammatory conditions in the tumor microenvironment promote angiogenesis, tumor progression, metastasis, and chemoresistance. Nuclear factor kappa B (NF-κB) regulates the expression of multiple pro-inflammatory genes and is therefore a key mediator of acute and chronic inflammatory responses.1, 2, 3, 4
Cancer stem cells (CSCs) possess the capacity for both self-renewal and differentiation; they promote tumor progression and metastasis and are responsible for chemoresistance and cancer relapse.5, 6, 7, 8 Accumulating evidence indicates that NF-κB activation supports the expansion of CSCs and promotes tumor development. The activation of NF-κB in cancer cells results in a number of downstream events, including induction of antiapoptotic genes and suppression of apoptosis. Moreover, induction of the regulators of epithelial–mesenchymal transition (EMT) leads to EMT phenotype, which has been hypothesized to initiate metastasis as well as the de-differentiation of cancer cells into CSCs.9, 10, 11, 12, 13 In contrast, cancer cells enriched for stemness have been recently shown to exhibit inflammatory gene expression patterns due to constitutive NF-κB activation;14, 15, 16, 17 this could lead to a positive feedback loop resulting in the amplification of inflammatory- and stemness-associated properties in cancer cells, although the molecular mechanism remains unclear.
Toll-like receptors (TLRs) are a group of proteins expressed by cells of the innate immune system that recognize microbial pathogens and respond to endogenous molecules released from dying cells during stress conditions such as chemotherapy.18, 19, 20 Interleukin (IL)-1 and tumor necrosis factor (TNF)-α are potent pro-inflammatory cytokines; TLR agonists, IL-1β, and TNF-α function as major pro-inflammatory stimuli in the tumor microenvironment.21, 22 Activation of TLRs and IL-1 receptor (IL-1R) triggers the sequential recruitment of MyD88, IRAK, and TRAF6 to form a complex; this, in turn, activates TAK, which leads to NF-κB activation. The activation of NF-κB downstream TNF-α receptor (TNFR) is mediated by the signaling molecules TRADD, RIP, and TRAF2. The molecular components involved in TLR/IL-1R and TNFR signaling pathways only partially overlap; nevertheless, the principle involved in the regulation of these pathways is similar and involves the recruitment of adaptor molecules, with protein levels and protein–protein interactions regulated by ubiquitination and deubiquitination. Several E3 ubiquitin-protein ligases, deubiquitinases, and co-factors involved in the ubiquitination system have been shown to regulate inflammatory properties of cancer cells through regulation of NF-κB activation.23, 24, 25, 26
MicroRNAs (or miRs) are a group of small (18–24 nt) non-coding RNAs that regulate target gene expression by binding target sites on the 3′ untranslated regions (UTRs) of messenger RNAs (mRNAs), leading to their degradation or translational inhibition.27, 28 Copper Metabolism MURR1 Domain-containing 1 (COMMD1) is the primary member of the COMM domain family that functions as an interface for protein–protein interactions and promotes ubiquitin-mediated degradation of the interaction partners.29, 30 The current study investigates the key molecules governing NF-κB activity and inflammatory properties in stemness-enriched cancer cells through analysis of microarray-derived expression profiles of negative regulators of the TLR, IL-1R, and TNFR signaling pathways in the course of stemness enrichment in head and neck squamous-cell carcinoma (HNSCC) cells. Parallel studies with HNSCC cells and non-small-cell lung cancer (NSCLC) cells indicated that upregulation of NF-κB-driven miR-205 in stemness-enriched cancer cells leads to COMMD1 downregulation; this results in amplification of inflammatory- and stemness-associated properties of cancer cells and also promotes in vivo tumorigenesis and tumor growth.
Results
Elevated NF-κB activation and inflammation in stemness-enriched cancer cells
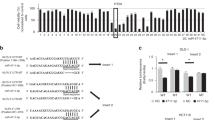
The changes in NF-κB activation and inflammation during the enrichment of stemness in cancer cells were investigated by analyzing microarray data derived from SAS HNSCC cells subject to enrichment for stemness in defined serum-free (DSF) selection medium for 2, 3, 5, or 9 weeks; this microarray data set was previously published31 and submitted to Gene Expression Omnibus (GEO) database (Accession No. GSE35603). Multiple NF-κB-regulated cytokines and chemokines were upregulated during the enrichment of stemness of SAS cells (Figure 1a), suggesting the elevation of NF-κB activity and inflammation during enrichment for stemness.
In silico identification of regulator of NF-κB activation during enrichment for stemness. (a) Elevated expression of NF-κB-regulated inflammatory genes during enrichment for stemness of HNSCC cells. The expression of NF-κB-regulated inflammatory genes was analyzed using microarray data (GEO data sets, GSE35603) obtained from SAS HNSCC cells at 0, 2, 3, 5, and 9 week time points during enrichment for stemness. (b) Inhibition of NF-κB activation by different regulators belonging to TNF-α, IL-1, and TLR signaling pathways. HEK 293 cells were co-transfected with NF-κB-regulated luciferase reporter plasmid and expression vector for the regulator in the absence (top and middle panels) or presence (bottom panel) of TLR2 expression vector. Cells were treated with TNF-α (10 ng/ml), IL-1 (10 ng/ml), or Pam3Cys (0.2 μg/ml) as indicated, and relative luciferase activities were determined. Data represent mean±S.D. from three independent experiments. *P<0.05; **P<0.01. (c) Heat map illustrating the expression profiles of negative regulators during enrichment of stemness in SAS HNSCC cells. (d) Reverse correlation between upregulation of NF-κB-controlled inflammatory genes (mean value from different probes of the gene) and downregulation of COMMD1 by Statistical Package for the Social Sciences spearman correlation analysis. (e) Expression levels of COMMD1 in normal and different types of tumors from patients. Data and P-values are provided by the Oncomine database
Identification of key regulators of NF-κB activation during enrichment for stemness
We hypothesized that downregulating the negative regulators of NF-κB activation in the TLR, IL-1R, and TNFR signaling pathways could lead to elevated NF-κB activation and inflammation in the stemness-enriched cancer cells. In order to identify the key regulator, a set of known negative regulators of NF-κB activation associated with these pathways were shortlisted for further analysis; these function through regulation of ubiquitination and deubiquitination processes. The inhibitory activities of these regulators on NF-κB activation induced by the TLR2 agonist Pam3Cys, IL-1β, and TNF-α were first confirmed by NF-κB luciferase reporter assay using TLR2-overexpressing or parental HEK 293 cells (Figure 1b). The expression profile of these regulators was also analyzed using the microarray data set obtained from stemness-enriched SAS cells in Figure 1a. Heat map generated from the microarray data revealed downregulation of a few of these negative regulators during the enrichment of stemness in SAS cells (Figure 1c). Of these, COMMD1 was the most downregulated gene with a good reverse correlation (R=−0.51) between its downregulation and the upregulated, NF-κB-controlled genes, including PTGS2, IL8, IL1A, CXCL2, IL-6, STAT5B, STAT3, CCL2, IL1B, CD40, and IL-15 (Figures 1a, c, and d). In addition, a search of the the Oncomine database revealed reduced expression of COMMD1 in different types of tumors from patients (Figure 1e). Thus, the functions of COMMD1 in the regulation of inflammatory- and stemness-associated properties of cancer cells were further investigated in this study.
Downregulation of COMMD1 in stemness-enriched HNSCC cells and NSCLC cells
The downregulation of COMMD1 observed from microarray data was further confirmed using SAS cells and also the human H460 and mouse D121 NSCLC cell lines. The cells were cultured in DSF medium to allow enrichment of sphere-forming cells exhibiting stemness and spheres were microscopically observed after 2 weeks (Figures 2a–c; top left panels). Real-time quantitative PCR (RT-qPCR) analysis revealed that COMMD1 expression was downregulated in the stemness-enriched SAS cells (Figure 2a; top right panel), which is consistent with microarray analysis (Figure 1c); similar observations were made in stemness-enriched H460 and D121 cells (Figures 2b and c; top right panels). RT-qPCR was employed for analyzing the expression of genes associated with stemness. Although the induction patterns of these genes differed between the three cell types, increased expression of these genes was observed in spheres compared with parental cells (Figures 2a–c; bottom panels), indicating the enrichment of stemness in cells of the sphere.
Downregulation of COMMD1 in stemness-enriched cancer cells. COMMD1 expression in parental and stemness-enriched (a) SAS, (b) H460, and (c) D121 cells. Top panels: cellular morphology of parental cells and stemness-enriched spheres (left) and corresponding COMMD1 expression (right). Bottom panels: expression of stemness-associated genes in parental cells and stemness-enriched spheres. Gene expression was analyzed by RT-qPCR. Data represent mean±S.D. from three independent experiments. *P<0.05; **P<0.01
3′-UTR of COMMD1 mRNA contains miR-205 target site
MicroRNAs have been implicated in the regulation of stemness in cancer cells through downregulation of their target genes.32, 33 To investigate the molecular mechanism underlying the downregulation of COMMD1, the 3′-UTR of COMMD1 mRNA was examined for microRNA target sites using the mirSVR scoring method.34, 35 Three target sites corresponding to miR-205, miR-491-5p, and miR-7, respectively, were predicted (Supplementary Figure S1a); of these, the miR-205 target site exhibited good mirSVR score of −1.2126 (Supplementary Figure S1b), which falls within the top 7% of mirSVR scores.35 Therefore, the regulation of COMMD1 expression by miR-205 was further investigated.
Downregulation of COMMD1 by NF-κB-regulated miR-205 during enrichment for stemness
To investigate whether miR-205 is responsible for COMMD1 downregulation in cancer cells enriched for stemness, the expression of miR-205 and COMMD1 were compared between parental cells and spheres cultured in DSF medium for 4 weeks through RT-qPCR; increased expression of miR-205 and downregulation of COMMD1 was observed in spheres derived from SAS, H460, and D121 cells (Figure 3a). Subsequent experiments aimed to investigate whether COMMD1 is a bona fide target of miR-205. Reporter constructs expressing luciferase regulated by the wild-type (wt) 3′-UTR of COMMD1 or a mutant (mut) containing mutated miR-205 target site were generated (Supplementary Figure S2a). These reporter constructs were co-transfected with miR-control or miR-205 mimic into HEK 293 cells stably expressing the control-sponge or miR-205-sponge designed to inhibit miR-205 (Supplementary Figure S2b). The experiment revealed that miR-205 destabilized the luciferase activity regulated by COMMD1 3′UTR-wt; this effect of miR-205 was antagonized by miR-205-sponge (Figure 3b, left panel). In contrast, miR-205 failed to exert any effect on luciferase activity regulated by COMMD1 3′-UTR-mut containing the mutant miR-205 target site (Figure 3b, right panel). Further studies revealed that overexpression of miR-205 resulting in COMMD1 downregulation at both the mRNA and protein levels in SAS, H460, and D121 cells (Figures 3c and d). A previous study reported the presence of NF-κB-binding site upstream to the miR-205 gene locus.36 Upregulation of miR-205 through NF-κB activation in NSCLC cells such as H460 has been reported to result in increased tumor growth.36 Thus, the induction of miR-205 in SAS and D121 cells was further investigated in the present study. MiR-205 was induced by IL-1β and TLR2 ligands, and the induction was reversed upon treatment of cells with BMS-345541, an IKK inhibitor37 (Figure 3e). Taken together, these results suggest that COMMD1 expression is downregulated by miR-205, which is upregulated by NF-κB activation during enrichment for stemness.
Downregulation of COMMD1 by NF-κB-regulated miR-205 in stemness-enriched cancer cells. (a) RT-qPCR analysis of miR-205 and COMMD1 expression in various cancer cell lines and stemness-enriched spheres. (b) HEK 293 cells stably expressing control or miR-205-sponge were co-transfected for 16 h with COMMD1 3′-UTR-wt or -mut luciferase reporter constructs and miR-control or miR-205 mimic, as indicated. Relative luciferase activities were determined. (c, d) Expression of COMMD1 in cells transfected with miR-control or miR-205 mimic was analyzed at mRNA and protein levels by RT-qPCR (c) and immunoblot (d), respectively. (e) Induction of miR-205 upon NF-κB activation. The cells were stimulated for 8 h with IL-1β (10 ng/ml) and Pam3Cys (0.2 μg/ml) in the presence or absence of BMS-345541. Expression of miR-205 primary transcript was determined by RT-qPCR. Data represent mean±S.D. from three independent experiments. *P<0.05; **P<0.01
COMMD1 downregulation in cancer cells promotes inflammation
The function of COMMD1 in regulating the inflammatory properties of cancer cells was investigated. To this end, shRNA viral vectors for the knockdown of COMMD1 mRNA (shCOMMD1) and luciferase mRNA (shLuc) were generated (Supplementary Figure 3). The control and COMMD1-knockdown cells were stimulated with TNF-α, IL-1, and various TLR ligands, and TNF-α expression was analyzed using RT-qPCR. COMMD1 knockdown was found to increase the responsiveness of SAS, H460, and D121 cells to inflammatory stimuli (Figures 4a–c). In RAW264.7 cells, a mouse monocytic cell line containing multiple TLRs, COMMD1 overexpression reversed NF-κB activation induced by TNF-α, IL-1, and various TLR ligands (Figure 4d), indicating that COMMD1 is a negative regulator of inflammatory responses induced by TNF-α, IL-1, and TLR ligands in monocytic and cancer cells.
COMMD1 regulates NF-κB activation upon inflammatory stimulation by promoting RelA downregulation. (a–d) shLuc and shCOMMD1 stably transfected SAS cells (a), H460 (b), D121 cells (c), and COMMD1 expression and NF-κB-regulated luciferase reporter vectors co-transfected RAW264.3 cells (d) were treated with vehicle control, TNF-α and IL-1 (10 ng/ml each), Pam3Cys, LPS, and flagellin (0.2 μg/ml each), polyI:C (2 μg/ml), as well as R848 and CpG-2006 (2 μ M each). Subsequent to cell lysis, NF-κB activation was determined in RAW264.3 cells by luciferase assay and in SAS, H460, and D121 cells by RT-qPCR for monitoring TNF-α induction. (e, f) HEK 293 cells were co-transfected for 16 h with expression vectors for COMMD1 and various signaling molecules of TNF-α, IL-1, and TLR pathways that regulate NF-κB activation as schematically illustrated in (f), and relative luciferase activities were determined (e). Data represent mean±S.D. from three independent experiments. *P<0.05; **P<0.01. (g) HEK 293 cells were co-transfected with expression vectors for COMMD1 and RelA, IKKβ, or TAK1, and protein levels were assessed by immunoblot analysis. (h) Expression levels of endogenous RelA in shLuc and shCOMMD1 stably transfected SAS, H460, and D121 cells. RelA was detected by immunoblot. Representative blots from three independent experiments are shown. Numbers on top of these blots show relative intensity of RelA expression in the respective shLuc and shCOMMD1 cells
The molecular target that mediates the regulation of inflammation by COMMD1 was investigated. The overexpression of various signaling molecules downstream TNFR, IL-1R, and TLR could cause NF-κB activation.38 COMMD1 was found to reverse NF-κB activation induced by these signaling molecules (Figures 4e and f), suggesting that the molecular target of COMMD1 is likely to be present further downstream in these pathways. Therefore, expression vectors for three downstream signaling molecules in the NF-κB activation pathways, namely, TAK1, IKKβ, and RelA, were co-transfected into HEK 293 cells in the presence or absence of COMMD1-expression vector; analysis of the expression of these three signaling molecules by immunoblot analysis revealed downregulation of RelA but not IKKβ and TAK1 upon COMMD1 co-expression (Figure 4g). Further, elevated expression levels of endogenous RelA were detected when COMMD1 was knocked down in SAS, H460, and D121 cells (Figure 4h), and increased phosphorylation levels of RelA (phospho-RelA), an indicator of NF-κB activation,39, 40 were measured by flow cytometric analysis in COMMD1-knockdown D121 cells (Supplementary Figure S4). These results suggest that RelA is the molecular target of COMMD1 in NF-κB activation pathways.
Downregulation of COMMD1 in cancer cells promotes macrophage recruitment
Subsequent investigation focused on the role of COMMD1 in regulating inflammation in the tumor microenvironment to modulate macrophage recruitment. Control and COMMD1-downregulated SAS and D121 cells were stimulated with TNF-α and the production of various cytokines and chemokines was analyzed by RT-qPCR using gene-specific primers (Supplementary Tables S2 and S3). Elevated levels of various cytokines and chemokines were observed in COMMD1-downregulated cells irrespective of TNF-α treatment (Figure 5a), suggesting that COMMD1 regulates both intrinsic and induced inflammatory responses in cancer cells. The role of COMMD1 in regulating the crosstalk between cancer cells and macrophages was further investigated through in vitro macrophage recruitment assay. Consistent with the production of various cytokines and chemokines by COMMD1-knockdown cells, the conditioned medium obtained from these cells proved more effective in macrophage recruitment (Figure 5b).
COMMD1 downregulation enhances intrinsic and TNF-α-induced inflammatory response and promotes macrophage recruitment. SAS and D121 cells stably transfected with shLuc or shCOMMD1 were treated with TNF-α (10 ng/ml) or control. (a) Induction of various cytokines was determined by RT-qPCR. (b) Conditioned medium collected from SAS (top panel) and D121 (bottom panel) cells from (a) was employed for assessing migration of THP-1 (top panel) and RAW264.3 (bottom panel) cells, respectively, in macrophage recruitment assay. In which, conditioned medium was placed in the lower chamber of transwell plates, while human monocytic THP-1 and mouse RAW264.7 cells were placed onto the upper chamber. Following incubation, the count of infiltrating macrophages was determined. Data represent mean±S.D. from three independent experiments. *,†P<0.05; **,††P<0.01
COMMD1 downregulation enhances stemness in cancer cells
The function of COMMD1 in regulating stemness was investigated. Analysis of the expression of stemness-associated genes using RT-qPCR revealed elevated expression of POU5F1, KLF4, NANOG, CD117, CD133, ALDH, and ABCG2 to varying extents in COMMD1-knockdown SAS and D121 cells compared with the respective controls (Figures 6a and b, left panels). The sphere-forming ability of COMMD1-knockdown cells was investigated in DSF medium. Decreased COMMD1 expression in cells correlated with increased number of spheres (Figures 6a and b, middle and right panels). Whether the function of COMMD1 was involved with NF-κB activation was further investigated and compared with the function of miR-205. In these experiments, different cell lines derived from H460 cells were treated with BMS-345541. Results indicated that knockdown of COMMD1 and overexpression of miR-205 increased the expression of stemness-associated genes and sphere formation, and these effects were blocked by the inhibition of NF-κB activation with BMS-345541 (Figures 6c and d). These results suggest that COMMD1 is a negative regulator and miR-205 is a positive regulator of stemness enrichment in cancer cells and that their function is mediated by NF-κB activation.
COMMD1 downregulation enhances stemness in cancer cells. (a) SAS and (b) D121 cells were stably transfected with shLuc or shCOMMD1. Expression of stemness-associated genes in control and COMMD1-knockdown cells was analyzed by RT-qPCR (left panels). Cells were cultured in DSF medium to obtain stemness-enriched sphere cells. Middle panels: representative photographs of spheres obtained. Right panels: enumeration of spheres. (c) H460 cells were stably transfected with shLuc or shCOMMD1. (d) H460 cells were stably transfected with control viral vector or viral vector for miR-205 overexpression. These cells were grown in the absence or presence of BMS-345541 (5 μ M). The expression of stemness-associated genes in control, COMMD1-knockdown, and miR-205 overexpression cells was analyzed (left panels). Number of spheres grown in DSF medium was counted (right panels). Data represent mean±S.D. from three independent experiments. *,†P<0.05; **,††P<0.01
COMMD1 downregulation enhances the transforming ability of cancer cells
The role of COMMD1 knockdown in regulating the transforming ability of cancer cells was investigated in parallel with the function of miR-205 overexpression using anchorage-independent growth assay. The control and COMMD1-knockdown cells of D121 (serial diluted from 2 × 104), H460 and SAS (1 × 104), and the control and miR-205-overexpressing cells of D121 and H460 (1 × 104) were cultured in soft agar for 2–3 weeks and colonies were enumerated. Significantly higher colony numbers were observed with COMMD1-knockdown cells and miR-205-overexpressing cells compared with the respective controls (Figures 7a and e). Thus, COMMD1 downregulation and miR-205 overexpression in cancer cells enhances their transforming ability, which is in agreement with the enhanced stemness associated with COMMD1 downregulation and miR-205 overexpression.
COMMD1 downregulation enhances colony formation in cancer cells. (a) Serial diluted cells of shLuc and shCOMMD1 stably transfected D121, and 1 × 104 of shLuc and shCOMMD1 stably transfected (b) H460 and (c) SAS cells, and 1 × 104 of control and miR-205 overexpression viral vector (pCDH) stably transfected (d) D121 and (e) H460 cells were growed in agarose-containing medium and colonies were enumerated after 2–3 weeks. Left panels: representative plates showing colony formation. Right bar figures: enumeration of colonies. Data represent mean±S.D. from three independent experiments. *P<0.05; **P<0.01
COMMD1 downregulation promotes in vivo tumorigenicity and tumor growth
The in vivo role of COMMD1 in regulation of tumorigenicity and tumor growth were investigated. C57BL/6J (B6) mice were inoculated with varying numbers of control or COMMD1-knockdown D121 cells. A higher tumor development rate was observed in mice injected with COMMD1-knockdown cells than in mice injected with control cells (Figure 8a). Tumor growth was investigated by inoculating (1 x 105) cells per mouse of COMMD1 knockdown, miR-205 overexpressing, and their respective control D121 cells; faster growth rates were observed in tumors derived from COMMD1-knockdown and miR-205-overexpressing cells relative to their control cells (Figures 8b and c). These observations suggest that downregulation of COMMD1 by miR-205 in cancer cells can promote tumorigenicity and tumor growth. The properties associated with inflammation and stemness were investigated in the tumors derived from COMMD1-knockdown and control cells. The expression of genes associated with inflammation and stemness was investigated in these tumors by RT-qPCR; higher expression of inflammatory cytokines and chemokines (Figure 8d) as well as stemness-associated genes (Figure 8e) was observed in tumors derived from COMMD1-knockdown cells. H&E staining of tumor sections revealed higher leukocyte infiltration in the tumors (Figure 8f). Moreover, flow cytometric analysis revealed an elevated level of phospho-RelA in whole tumor cells, Cd11b+ tumor-associated leukocytes, and Cd117+ stemness-enriched tumor cells in tumors derived from COMMD1-knockdown cells relative to their respective cells in tumors derived from control cells (Figures 8g and i). Flow cytometric analysis also showed expanded populations of Cd11b+ leukocytes and Cd117+ stemness-enriched cells in the tumors derived from COMMD1-knockdown cells (Figures 8h and i). The Cd117+ cells and Cd117− were isolated from the tumors grown from COMMD1-knockdown cells and reinjected into mice to access their capacity for tumorigenesis. Results showed more potency for the Cd117+ cells to develop tumors than the Cd117− cells (Figure 8j), confirming that the Cd117+ stemness-enriched cells were more aggressive cancer cells. Taken together, these observations suggest that COMMD1 regulates tumorigenicity and tumor growth by modulating inflammatory- and stemness-associated properties of cancer cells.
COMMD1 downregulation promotes tumorigenesis and tumor growth. (a) C57BL/6 mice were subcutaneously injected with varying counts of D121 cells stably transfected with shCOMMD1 or shLuc, and mice exhibiting tumor growth were enumerated 28 days post-injection. (b) D121 cells (1 × 105) stably transfected with shCOMMD1 or shLuc, and (c) D121 cells (1 × 105) stably transfected with control or miR-205 overexpression viral vector were subcutaneously injected into C57BL/6 mice. Tumor volume was measured at the indicated time points, and mean tumor size was plotted (mean±S.D., n=6). Tumor tissues were collected 20 days post-injection. The expression of (d) inflammatory and (e) stemness-associated genes was analyzed by RT-qPCR, and (f) tissue sections were stained with H&E for detecting leukocyte infiltration. Tumor cells were suspended in PBS and stained for different markers, the cell population and the level of phospho-RelA in different cells were analyzed using flow cytometry. (g) The left panel shows a representative histogram for the right bar figure for phospho-RelA levels in the whole tumor cells from shLuc and shCOMMD1 stably transfected cells derived tumors. (h, i) The left two panels show a set of representative histograms for the middle bar figure for analysis population of the Cd11b+ tumor-associated leukocytes (h) and Cd117+ stemness-enriched tumor cells (i) in whole tumor cells. The forth panel shows a representative histogram for the right bar figure for phospho-RelA levels in the indicated cell population. Data represent mean±S.D. from three independent experiments. *P<0.05; **P<0.01. (j) C57BL/6 mice were subcutaneously injected with varying counts of Cd117+ and Cd117− tumor cells, and mice exhibiting tumor growth were enumerated 28 days post-injection
Discussion
Tumor-promoting inflammation has been recognized as a hallmark of cancer.1 NF-κB is key mediator of inflammation in cancer cells.9, 10, 11 CSCs are implicated in tumor recurrence, metastasis, and higher mortality rates.7, 8 Recent studies have supported a role for NF-κB in modulation of stemness in cancer cells and in inflammatory signaling, which is important for CSC self-renewal and maintenance.12, 13 Moreover, pro-inflammatory cytokines and TLR agonists in inflammatory tumor microenvironment support the survival and tumorigenic activities of CSCs, whereas NF-κB inhibitors have been shown to inhibit CSC growth.12, 13, 41, 42, 43, 44 These observations suggest that the mechanism underlying the promotion of malignancy by inflammation could involve enhancement of stemness in cancer cells resulting in increased CSC population. Apart from these, the present and several previous studies14, 15, 16, 17 revealed increased expression of inflammatory genes in CSCs due to constitutive NF-κB activation; this can further promote the release of pro-inflammatory mediators into the tumor microenvironment, thereby forming a positive feedback loop for increasing stemness in cancer cells and amplifying inflammation in the cancer cells, CSCs, and the tumor microenvironment. This positive feedback loop is likely to be an important mechanism for malignancy, despite the lack of clarity regarding the underlying molecular controls.
TLRs, IL-1R, and TNFR initiate potent pro-inflammatory signaling pathways resulting in NF-κB activation in cancer cells, thereby promoting cancer cell growth, CSC expansion, and tumor development.21, 22 These signaling pathways are regulated by multiple negative regulators that mediate ubiquitination and deubiquitination of signaling molecules to modulate protein–protein interactions and proteasomal degradation.23, 24, 25, 26 These negative regulators not only modulate the magnitude of inflammatory responses to stimuli but also maintain the balance of NF-κB activity within cells. Thus, downregulation of these negative regulators is likely to cause the elevated NF-κB activation observed during enrichment of stemness in cancer cells and in the CSCs. In this study, our results reveal that miR-205-mediated downregulation of COMMD1 is responsible for the increased inflammatory and stemness features in stemness-enriched cancer cells and promotes tumorigenesis and tumor growth.
Both antitumor and protumor functions of miR-205 have been reported.45, 46 The antitumor activity of miR-205 is attributable to its targeting various oncogenes. For example, expression of miR-205 suppresses cell proliferation by suppressing E2F1 expression in melanoma cells, and by inhibition of Src-mediated oncogenic pathways for its growth-inhibitory effects in renal carcinoma.47, 48 In breast cancer cells, knockdown of miR-205 enhances stem cell traits and promotes development of mammary lesions in cancer animal model.49 In addition, miR-205 is downregulated in gastric cancer and miR-205 inhibition promotes the proliferation of gastric cancer cells.50 On the other hand, accumulating evidence indicates that miR-205 promotes tumorigenesis, tumor progression, and chemoresistance. In mouse mammary epithelial cells, miR-205 expression is enhanced in stem-like cells and suppresses phosphatase and tensin homolog (PTEN), leading to the expansion of progenitor cell population.51 In human breast cancer cells, miR-205 overexpression increases the enrichment of stem-like cells exhibiting self-renewal ability.52 Elevated expression of miR-205 is associated with poor prognosis in endometrial tumors, with miR-205 promoting the proliferation of endometrial cancer cells.53 In addition, miR-205 expression correlates with the initiation of nasopharyngeal carcinoma and contributes to its radioresistance.54 In ovarian cancer cells, miR-205 is upregulated upon VEGF-A treatment and targets Ezrin and Lamin A/C to promote invasion and proliferation.55 Furthermore, several studies have shown that upregulation of miR-205 in NSCLC cells promotes growth, invasion, and chemoresistance of the cancer cells.36, 56, 57 These observations suggest that the pro- or antitumor functions of miR-205 are determined by its target genes, the context of cancer, and the tumor microenvironment.
In contrast, COMMD1 is shown to have an antitumor function. Previous studies suggested that COMMD1 inhibits NF-κB activation by functioning as a scaffold protein in conjugation with ECSSOCS1, a multimeric E3 ubiquitin ligase complex, to promote ubiquitination and proteasomal degradation of the NF-κB subunit RelA.58 The conditional knockdown of COMMD1 in myeloid cells rendered mice hyper-responsive to LPS stimulation as well as more sensitive to dextran sodium sulfate-induced colitis and colitis-associated cancer.59 In concordance with these results, reduced COMMD1 expression was observed in several human cancers and low expression of COMMD1 correlated with a reduced survival rate among patients with endometrial cancer.60 The level of COMMD1 protein is negatively regulated by secretory clusterin (sCLU), a stress-induced small heat-shock protein, in prostate cancer cells. The expression of sCLU showed inverse correlation with that of COMMD1 but positive correlation with NF-κB-regulated genes.61 Consistent with this observation, the present study showed that COMMD1 downregulation promotes tumorigenesis and tumor growth. Downregulation of COMMD1 during the enrichment of stemness in cancer cells resulted in upregulation of genes associated with inflammation and stemness. Furthermore, the present study shows that COMMD1 downregulation can be regulated at the level of mRNA stability by the NF-κB-regulated miR-205, in addition to the previous observed regulation at protein level by sCLU.61
In summary, this study has shown that during the enrichment of stemness in HNSCC cells and NSCLC cells, COMMD1 is downregulated by miR-205, while miR-205 is upregulated upon NF-κB activation. The downregulation of COMMD1 promotes the expression of stemness-associated genes and enhances inflammation-induced as well as sustained intrinsic NF-κB activation in cancer cells. This, in turn, promotes the release of pro-inflammatory mediators into the tumor microenvironment and further enhances miR-205 expression in cancer cells. Thus, COMMD1 plays a role in the regulation of tumorigenesis and tumor growth via regulation of a positive feedback loop that amplifies inflammatory- and stemness-associated properties in cancer cells.
Materials and Methods
Reagents and antibodies
The TLR agonists Pam3Cys, polyI:C, LPS, flagellin, and R848 were purchased from Invivogen (San Diego, CA, USA); CpG-2006 and CpG-1826 from Invitrogen (Carlsbad, CA, USA) or Genomics Biosci & Tech (New Taipei, Taiwan); BMS-345541, collagenase II, collagenase IV, deoxyribonuclease, and anti-Flag M2 antibody from Sigma-Aldrich Co. (St. Louis, MO, USA); and HA.11, PE-anti-human CD117, PE- anti-mouse CD117, and PE-rat IgG2b κ isotype control antibody from BioLegend (San Diego, CA, USA); PE-anti-human CD133 antibody from Miltenyi Biotec (Bergisch Gladbach, Germany); PE-anti-mouse CD11b antibody from eBioscience (San Diego, CA, USA); anti-phospho-RelA (Alexa Fluor 647 conjugate) and rabbit IgG isotype control antibody from Cell Signaling (Beverley, MA, USA). The miR-205 mimics and control were synthesized by GeneDirex (Gueishan Township, Taiwan). Antibodies against human and mouse COMMD1 were purchased from Abnova (Taipei, Taiwan) and Proteintech (Chicago, IL, USA), respectively; human recombinant TNF-α, IL-1β, epidermal growth factor (EGF), and basic fibroblast growth factor (bFGF) from Peprotech (Rocky Hill, NJ, USA); and reagents for luciferase assay, from Promega (Medison, WI, USA).
Cell culture and enrichment of sphere-forming cancer cells
Human SAS HNSCC, HEK 293, murine D121 NSCLC, and RAW264.7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Human H460 NSCLC and THP-1 cells were grown in RPMI supplemented with 10% FBS. For enrichment of sphere-forming cells, SAS, H460, and D121 cells were cultured at a density of 10 000 cells/ml in six-well plates coated with poly(2-hydroxyethyl methacrylate) (polyHEMA; Sigma-Aldrich Co.) in sphere medium consisting of serum-free DMEM/F12-K medium, N2 supplement (GIBCO, Carlsbad, CA, USA), 20 ng/ml human recombinant EGF, and 20 ng/ml human recombinant bFGF. PolyHEMA-coated plates were prepared by dissolving 10 mg/ml polyHEMA in 95% ethanol; 1 ml of the solution was added onto the six-well plates and placed overnight in a laminar flow hood at room temperature for air drying.
Bioinformatics analysis of microarray data, gene expression in tumor, and identification of miRNA target sites
Microarray data obtained at various time points during the enrichment of stemness for SAS HNSCC cells (GEO data sets, GSE35603) were analyzed for expression profiles of NF-κB-regulated inflammatory genes as well as negative regulators of the TLR, IL-1R, and TNFR signaling pathways. Oncomine database (https://www.oncomine.org/) was searched for the expression profiles of COMMD1 in normal tissues and tumors from patients. MicroRNA target sites in the 3′-UTR of COMMD1 mRNA were analyzed using the algorithm miRanda (http://www.microrna.org/).
Plasmid construction
The expression constructs for human COMMD1, PDLIM2, SOCS1, CYLD, USP4, USP7, USP25, and TRIM38 were generated through PCR amplification of the corresponding protein-coding regions from the first-strand cDNA library derived from human spleen cells generated as previous reported.62 The amplified DNA fragments were cloned into pRK5 vector for protein expression. The 3′-UTR reporter constructs were generated through PCR amplification of the 3′-UTR of COMMD1 from human genomic DNA purchased from Clontech Laboratories, Inc. (Mountain View, CA, USA), followed by cloning into pMIR-REPORT luciferase vector.
Lentiviral shRNA, miR-205-sponge and precursor miR-205 construction and infection
The shRNA constructs were generated by synthesizing and annealing sense and antisense shRNA oligonucleotides with 5′GATC and 5′AATT overhangs, respectively. The double-stranded shRNA was cloned into the BamHI/EcoRI sites of pGreenpuro plasmid. The miR-205-sponge was constructed by annealing sense and antisense oligonucleotides containing six tandem repeats of the bulge-containing miR-205-binding motif and cloning into XbaI/BamHI sites of pCDH plasmid. Precursor miR-205 construct was generated through PCR amplification of the precursor miR-205 from human genomic DNA, followed by cloning into EcoRI/BamHI sites of pCDH plasmid. Lentivirus was produced by harvesting culture supernatants obtained upon transfecting 293 T cells with the generated constructs along with three packaging plasmids. HEK 293 and cancer cell lines were spin-infected by plating cells in 12-well plates in the presence of lentiviral supernatants and 8 μg/ml polybrene (Sigma-Aldrich Co.), followed by centrifugation at 1100 × g for 30 min. The cells were subject to selection with puromycin (3 ng/ml) to obtain stable cell lines. The sequences of oligonucleotides employed for generating shRNA, miR-205, and miR-205-sponge constructs, and precursor miR-205 are listed (Supplementary Tables S1).
Luciferase reporter assays
For NF-κB activation assay, cells were plated on 24-well plates, allowed to adhere overnight, and co-transfected with NF-κB-driven luciferase reporter, β-galactosidase, and the indicated expression plasmids using polyethylenimine (Sigma-Aldrich Co.). The following day, the cells were subject to 10-h treatment with TNF-α (10 ng/ml), IL-1β (10 ng/ml), or Pam3Cys (200 ng/ml), followed by lysis and determination of luciferase activity. Relative luciferase activities were calculated as fold induction relative to unstimulated control. For 3′-UTR reporter assay, HEK 293 cells stably transfected with control or miR-205-sponge were co-transfected for 16 h with COMMD1 3′-UTR luciferase reporter plasmid, β-galactosidase plasmid, and the indicated microRNA precursors, followed by cell lysis and determination of luciferase activities.
SDS-PAGE and immunoblot analysis
Cells were lysed with lysis buffer (100 mM NaCl, 50 mM Tris-Cl pH 7.5, 0.5 mM EDTA, 1% NP-40) containing complete protease inhibitor cocktail (Roche Life Science, Indianapolis, IN, USA). Cell lysates were separated by SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked for 30 min with 5% fat-free milk in Tris-buffered saline containing Tween-20 (TBST; 50 mM Tris-Cl pH 7.5, 150 mM NaCl, 0.1% Tween-20) and incubated for 2 h with the indicated antibody in TBST containing 2% fat-free milk. The membranes were subsequently washed in TBST and incubated for 1 h with HRP-conjugated secondary antibody. The immunoreactive bands were visualized using chemiluminescent HRP substrate (Immobilon Western; Millipore, Temecula, CA, USA) and the UVP BioSpectrum Imaging System.
RT-qPCR analysis of gene and microRNA expression
Total RNA including small RNA species was purified using TRIzol (Invitrogen), according to the manufacturer’s instructions. Reverse transcription (RT) was performed using SuperScript III first-strand synthesis system (Invitrogen) and oligo-dT for first-strand cDNA synthesis or corresponding RT primers for miRNA synthesis. RT-qPCR was carried out using ABI PRISM 7900HT Sequence Detection System and KAPA SYBR fast qPCR kit (KK4605) for gene expression analysis or FastStart Universal Probe Master and Universal ProbeLibrary Probe #21 (Roche Life Science) for microRNA detection. Data were analyzed using the 2−ΔΔCt method described in the ABI user manual. The expression of mRNA and microRNA were normalized to β-actin and RNU6, respectively. The primer sequences are listed (Supplementary Table S1–S3).
Anchorage-independent growth
Bottom agar–medium mixture (DMEM, 10% FBS, 0.8% agarose) was added to each well of six-well tissue culture plates and allowed to solidify. Top agar–medium mixture (DMEM, 10% FBS, 0.4% agarose) containing (1 × 104) cells was added, followed by the addition of growth medium. The plates were incubated at 37 °C for 2–3 weeks and colony formation was monitored. The plates were fixed with 4% formaldehyde and stained with 0.005% crystal violet in PBS for 1 h. The colonies were photographed and counted. The total number of colonies with a diameter of ≥100 μm were quantified over four fields per well for a total of 12 fields in triplicate samples.
Macrophage recruitment assay
Conditioned medium (CM) was collected from cancer cells treated with TNF-α or control for 24 h and added to the lower chamber of transwell plates containing polyethylene terephthalate (PET) membrane insert of 5-μm pore size (Corning Life Sciences, Manassas, VA, USA). THP-1 (1 × 105) or RAW264.7 (2 × 104) cells were plated onto the upper chamber and incubated for 8 h at 37 °C. Cells that failed to migrate through the pores were removed using a cotton swab, while the migratory cells were fixed, stained with 0.05% crystal violet, and counted in five randomly selected fields.
Flow cytometric analysis
For flow cytometric analysis, cells were suspended in PBS containing 2% FCS and incubated with PE-conjugated Abs as indicated at 4 °C for 30 min. After washing, cells were analyzed on a FACSCalibur flow cytometer with CellQuest software (Becton Dickinson, San Jose, CA, USA). For intracellular staining of phospho-RelA, cell line and tumor mass cells were first fixed and permeabilized by BD Cytofix/Cytoperm Kit (BD Bioscience, San Diego, CA, USA) for 20 min. After fixation and permeabilization, cells were washed and resuspensed in 1 × BD Perm/wash buffer and then incubated with anti-phospho-RelA for 30 min.
Animal care, preparation of tumor single-cell suspension, tumorigenesis, and analysis of tumor growth
Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the National Health Research Institutes, Taiwan. C57BL/6J mice were maintained and handled in accordance with the stated guidelines. D121 cells stably expressing shLuc and shCOMMD1 were harvested, resuspended in PBS, and injected subcutaneously into C57BL/6 mice of 6–8 weeks of age. Tumor single-cell suspension was prepared by mincing and digestion of tumor mass in PBS containing 0.5% BSA, 0.25% collagenase II, 0.25% collagenase IV, and 0.05% deoxyribonuclease for 30 min. The reaction was stopped with DMEM containing 10% FBS. The mixture was then strained through a 70- μ M strainer and lysis of red blood cells with RBC lysis buffer (eBioscience). Cd117+ tumor cells were separated using MACS from tumor single-cell suspension with Cd117 MicroBeads and MACS column (Miltenyi Biotec) following the manufacturer’s instruction. Tumor volume (TV) was calculated using the following formula: TV (mm3)=(length × width2)/2.
Histological analysis
Tissue samples were collected, immersed in 10% formalin, embedded in paraffin wax, and sectioned. The sections were stained with H&E and microscopically visualized.
Statistical analysis
Data are expressed as mean±S.D. Statistical analysis was performed on data derived from three or more independent experiments using Student’s t-test. A P-value of <0.05 was considered to represent statistically significant differences between the experimental groups.
Abbreviations
- COMMD1:
-
copper metabolism MURR1 domain-containing 1
- NF-κB:
-
nuclear factor-κB
- TLR:
-
toll-like receptor
- IL-1:
-
interleukin 1
- TNF-α:
-
tumor necrosis factor-α
- CSC:
-
cancer stem cells
- HNSCC:
-
head and neck squamous-cell carcinoma
- NSCLC:
-
non-small cell lung cancer
References
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Mantovani A, Allavena P, Sica A, Balkwill F . Cancer-related inflammation. Nature 2008; 454: 436–444.
Grivennikov SI, Greten FR, Karin M . Immunity, inflammation, and cancer. Cell 2010; 140: 883–899.
Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA . Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer 2013; 13: 759–771.
Reya T, Morrison SJ, Clarke MF, Weissman IL . Stem cells, cancer, and cancer stem cells. Nature 2001; 414: 105–111.
Visvader JE, Lindeman GJ . Cancer stem cells: current status and evolving complexities. Cell Stem Cell 2012; 10: 717–728.
Mertins SD . Cancer stem cells: a systems biology view of their role in prognosis and therapy. Anticancer Drugs 2014; 25: 353–367.
Colak S, Medema JP . Cancer stem cells – important players in tumor therapy resistance. FEBS J 2014; 281: 4779–4791.
Gasparini C, Feldmann M . NF-kappaB as a target for modulating inflammatory responses. Curr Pharm Des 2012; 18: 5735–5745.
Sarkar FH, Li Y, Wang Z, Kong D . NF-kappaB signaling pathway and its therapeutic implications in human diseases. Int Rev Immunol 2008; 27: 293–319.
DiDonato JA, Mercurio F, Karin M . NF-kappaB and the link between inflammation and cancer. Immunol Rev 2012; 246: 379–400.
Zhou C, Liu J, Tang Y, Liang X . Inflammation linking EMT and cancer stem cells. Oral Oncol 2012; 48: 1068–1075.
Shigdar S, Li Y, Bhattacharya S, O'Connor M, Pu C, Lin J et al. Inflammation and cancer stem cells. Cancer Lett 2014; 345: 271–278.
Rajasekhar VK, Studer L, Gerald W, Socci ND, Scher HI . Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-kappaB signalling. Nat Commun 2011; 2: 162.
Garner JM, Fan M, Yang CH, Du Z, Sims M, Davidoff AM et al. Constitutive activation of signal transducer and activator of transcription 3 (STAT3) and nuclear factor kappaB signaling in glioblastoma cancer stem cells regulates the Notch pathway. J Biol Chem 2013; 288: 26167–26176.
Yamashina T, Baghdadi M, Yoneda A, Kinoshita I, Suzu S, Dosaka-Akita H et al. Cancer stem-like cells derived from chemoresistant tumors have a unique capacity to prime tumorigenic myeloid cells. Cancer Res 2014; 74: 2698–2709.
Jinushi M . Role of cancer stem cell-associated inflammation in creating pro-inflammatory tumorigenic microenvironments. Oncoimmunology 2014; 3: e28862.
Hoebe K, Jiang Z, Georgel P, Tabeta K, Janssen E, Du X et al. TLR signaling pathways: opportunities for activation and blockade in pursuit of therapy. Curr Pharm Des 2006; 12: 4123–4134.
Chen K, Huang J, Gong W, Iribarren P, Dunlop NM, Wang JM . Toll-like receptors in inflammation, infection and cancer. Int Immunopharmacol 2007; 7: 1271–1285.
Piccinini AM, Midwood KS . DAMPening inflammation by modulating TLR signalling. Mediators Inflamm 2010; 2010 pii: 672395.
Lawrence T . The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol 2009; 1: a001651.
Hoesel B, Schmid JA . The complexity of NF-kappaB signaling in inflammation and cancer. Mol Cancer 2013; 12: 86.
Verstrepen L, Bekaert T, Chau TL, Tavernier J, Chariot A, Beyaert R . TLR-4, IL-1R and TNF-R signaling to NF-kappaB: variations on a common theme. Cell Mol Life Sci 2008; 65: 2964–2978.
Bibeau-Poirier A, Servant MJ . Roles of ubiquitination in pattern-recognition receptors and type I interferon receptor signaling. Cytokine 2008; 43: 359–367.
Wertz IE, Dixit VM . Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb Perspect Biol 2010; 2: a003350.
Zinngrebe J, Montinaro A, Peltzer N, Walczak H . Ubiquitin in the immune system. EMBO Rep 2014; 15: 28–45.
Bushati N, Cohen SM . microRNA functions. Annu Rev Cell Dev Biol 2007; 23: 175–205.
Huntzinger E, Izaurralde E . Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 2011; 12: 99–110.
Maine GN, Burstein E . COMMD proteins and the control of the NF kappa B pathway. Cell Cycle 2007; 6: 672–676.
Bartuzi P, Hofker MH, van de Sluis B . Tuning NF-kappaB activity: a touch of COMMD proteins. Biochim Biophys Acta 2013; 1832: 2315–2321.
Lo JF, Yu CC, Chiou SH, Huang CY, Jan CI, Lin SC et al. The epithelial-mesenchymal transition mediator S100A4 maintains cancer-initiating cells in head and neck cancers. Cancer Res 2011; 71: 1912–1923.
Iorio MV, Croce CM . microRNA involvement in human cancer. Carcinogenesis 2012; 33: 1126–1133.
Garofalo M, Croce CM . Role of microRNAs in maintaining cancer stem cells. Adv Drug Deliv Rev 2015; 81: 53–61.
Betel D, Wilson M, Gabow A, Marks DS, Sander C . The microRNA.org resource: targets and expression. Nucleic Acids Res 2008; 36: D149–D153.
Betel D, Koppal A, Agius P, Sander C, Leslie C . Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 2010; 11: R90.
Cai J, Fang L, Huang Y, Li R, Yuan J, Yang Y et al. miR-205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non-small cell lung cancer. Cancer Res 2013; 73: 5402–5415.
McIntyre KW, Shuster DJ, Gillooly KM, Dambach DM, Pattoli MA, Lu P et al. A highly selective inhibitor of I kappa B kinase, BMS-345541, blocks both joint inflammation and destruction in collagen-induced arthritis in mice. Arthritis Rheum 2003; 48: 2652–2659.
Xu C, Liu J, Hsu LC, Luo Y, Xiang R, Chuang TH . Functional interaction of heat shock protein 90 and Beclin 1 modulates Toll-like receptor-mediated autophagy. FASEB J 2011; 25: 2700–2710.
Park GY, Christman JW . Nuclear factor kappa B is a promising therapeutic target in inflammatory lung disease. Curr Drug Targets 2006; 7: 661–668.
Vallabhapurapu S, Karin M . Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol 2009; 27: 693–733.
Chefetz I, Alvero AB, Holmberg JC, Lebowitz N, Craveiro V, Yang-Hartwich Y et al. TLR2 enhances ovarian cancer stem cell self-renewal and promotes tumor repair and recurrence. Cell Cycle 2013; 12: 511–521.
Chen K, Huang YH, Chen JL . Understanding and targeting cancer stem cells: therapeutic implications and challenges. Acta Pharmacol Sin 2013; 34: 732–740.
Liu WT, Jing YY, Yu GF, Han ZP, Yu DD, Fan QM et al. Toll like receptor 4 facilitates invasion and migration as a cancer stem cell marker in hepatocellular carcinoma. Cancer Lett 2015; 358: 136–143.
Pistollato F, Giampieri F, Battino M . The use of plant-derived bioactive compounds to target cancer stem cells and modulate tumor microenvironment. Food Chem Toxicol 2015; 75: 58–70.
Qin AY, Zhang XW, Liu L, Yu JP, Li H, Wang SZ et al. MiR-205 in cancer: an angel or a devil? Eur J Cell Biol 2013; 92: 54–60.
Orang AV, Safaralizadeh R, Hosseinpour Feizi MA . Insights into the diverse roles of miR-205 in human cancers. Asian Pac J Cancer Prev 2014; 15: 577–583.
Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M . miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem 2011; 286: 16606–16614.
Majid S, Saini S, Dar AA, Hirata H, Shahryari V, Tanaka Y et al. MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal cancer. Cancer Res 2011; 71: 2611–2621.
Chao CH, Chang CC, Wu MJ, Ko HW, Wang D, Hung MC et al. MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J Clin Invest 2014; 124: 3093–3106.
Yin WZ, Li F, Zhang L, Ren XP, Zhang N, Wen JF . Down-regulation of microRNA-205 promotes gastric cancer cell proliferation. Eur Rev Med Pharmacol Sci 2014; 18: 1027–1032.
Greene SB, Gunaratne PH, Hammond SM, Rosen JM . A putative role for microRNA-205 in mammary epithelial cell progenitors. J Cell Sci 2010; 123: 606–618.
Vilquin P, Donini CF, Villedieu M, Grisard E, Corbo L, Bachelot T et al. MicroRNA-125b upregulation confers aromatase inhibitor resistance and is a novel marker of poor prognosis in breast cancer. Breast Cancer Res 2015; 17: 13.
Karaayvaz M, Zhang C, Liang S, Shroyer KR, Ju J . Prognostic significance of miR-205 in endometrial cancer. PLoS One 2012; 7: e35158.
Qu C, Liang Z, Huang J, Zhao R, Su C, Wang S et al. MiR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle 2012; 11: 785–796.
Li J, Li L, Li Z, Gong G, Chen P, Liu H et al. The role of miR-205 in the VEGF-mediated promotion of human ovarian cancer cell invasion. Gynecol Oncol 2015; 137: 125–133.
Lei L, Huang Y, Gong W . miR-205 promotes the growth, metastasis and chemoresistance of NSCLC cells by targeting PTEN. Oncol Rep 2013; 30: 2897–2902.
Larzabal L, de Aberasturi AL, Redrado M, Rueda P, Rodriguez MJ, Bodegas ME et al. TMPRSS4 regulates levels of integrin alpha5 in NSCLC through miR-205 activity to promote metastasis. Br J Cancer 2014; 110: 764–774.
Maine GN, Mao X, Komarck CM, Burstein E . COMMD1 promotes the ubiquitination of NF-kappaB subunits through a cullin-containing ubiquitin ligase. EMBO J 2007; 26: 436–447.
Li H, Chan L, Bartuzi P, Melton SD, Weber A, Ben-Shlomo S et al. Copper metabolism domain-containing 1 represses genes that promote inflammation and protects mice from colitis and colitis-associated cancer. Gastroenterology 2014; 147: e3.
van de Sluis B, Mao X, Zhai Y, Groot AJ, Vermeulen JF, van der Wall E et al. COMMD1 disrupts HIF-1alpha/beta dimerization and inhibits human tumor cell invasion. J Clin Invest 2010; 120: 2119–2130.
Zoubeidi A, Ettinger S, Beraldi E, Hadaschik B, Zardan A, Klomp LW et al. Clusterin facilitates COMMD1 and I-kappaB degradation to enhance NF-kappaB activity in prostate cancer cells. Mol Cancer Res 2010; 8: 119–130.
Liu J, Xu C, Hsu LC, Luo Y, Xiang R, Chuang TH . A five-amino-acid motif in the undefined region of the TLR8 ectodomain is required for species-specific ligand recognition. Mol Immunol 2010; 47: 1083–1090.
Acknowledgements
We thank the Laboratory Animal Center of the National Health Research Institutes, Taiwan, for assistance with animal work. This work was supported in part by the National Health Research Institutes, Taiwan (grant IM-103-PP-02) and Ministry of Science and Technology of Taiwan (grants MOST102-2320-B-400-009-MY3). D-WY carried out thesis research under the auspices of the Graduate Program of Biotechnology in Medicine, National Tsing-Hua University, and National Health Research Institutes.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by R De Maria
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Yeh, DW., Chen, YS., Lai, CY. et al. Downregulation of COMMD1 by miR-205 promotes a positive feedback loop for amplifying inflammatory- and stemness-associated properties of cancer cells. Cell Death Differ 23, 841–852 (2016). https://doi.org/10.1038/cdd.2015.147
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2015.147
This article is cited by
-
Long non-coding RNA SNHG5 suppresses the development of acute respiratory distress syndrome by targeting miR-205/COMMD1 axis
Molecular and Cellular Biochemistry (2021)
-
Porphyromonas gingivalis Inhibition of MicroRNA-205-5p Expression Modulates Proinflammatory Cytokines in Gingival Epithelial Cells
Biochemical Genetics (2020)
-
Prognosis and modulation mechanisms of COMMD6 in human tumours based on expression profiling and comprehensive bioinformatics analysis
British Journal of Cancer (2019)
-
USP17 mediates macrophage-promoted inflammation and stemness in lung cancer cells by regulating TRAF2/TRAF3 complex formation
Oncogene (2018)
-
MicroRNA-transcription factor network analysis reveals miRNAs cooperatively suppress RORA in oral squamous cell carcinoma
Oncogenesis (2018)