Abstract
Cleavage of eukaryotic translation initiation factor 4G (eIF4G) by enterovirus proteases during infection leads to the shutoff of cellular cap-dependent translation, but does not affect the initiation of cap-independent translation of mRNAs containing an internal ribosome entry site (IRES). Death-associated protein 5 (DAP5), a structural homolog of eIF4G, is a translation initiation factor specific for IRES-containing mRNAs. Coxsackievirus B3 (CVB3) is a positive single-stranded RNA virus and a primary causal agent of human myocarditis. Its RNA genome harbors an IRES within the 5′-untranslated region and is translated by a cap-independent, IRES-driven mechanism. Previously, we have shown that DAP5 is cleaved during CVB3 infection. However, the protease responsible for cleavage, cleavage site and effects on the translation of target genes during CVB3 infection have not been investigated. In the present study, we demonstrated that viral protease 2A but not 3C is responsible for DAP5 cleavage, generating 45- and 52-kDa N- (DAP5-N) and C-terminal (DAP5-C) fragments, respectively. By site-directed mutagenesis, we found that DAP5 is cleaved at amino acid G434. Upon cleavage, DAP5-N largely translocated to the nucleus at the later time points of infection, whereas the DAP5-C largely remained in the cytoplasm. Overexpression of these DAP5 truncates demonstrated that DAP5-N retained the capability of initiating IRES-driven translation of apoptosis-associated p53, but not the prosurvival Bcl-2 (B-cell lymphoma 2) when compared with the full-length DAP5. Similarly, DAP5-N expression promoted CVB3 replication and progeny release; on the other hand, DAP5-C exerted a dominant-negative effect on cap-dependent translation. Taken together, viral protease 2A-mediated cleavage of DAP5 results in the production of two truncates that exert differential effects on protein translation of the IRES-containing genes, leading to enhanced host cell death.
Similar content being viewed by others
Main
Coxsackievirus B3 (CVB3), a primary cause of viral myocarditis, is associated with sudden, unexpected death,1 dilated cardiomyopathy and heart failure.2 The CVB3 genome consists of a single open reading frame, which is translated to a polyprotein, and subsequently processed by viral proteases 2A and 3C. Similar to other picornaviruses, the CVB3 genome is linked to a small viral peptide, Vpg, rather than a 7-methyl guanosine cap structure at its 5′ terminus. Thus, CVB3 RNA is translated by a cap-independent mechanism and is driven by an internal ribosome entry site (IRES) harbored in the 5′ untranslated region (5′-UTR).3, 4
Viral proteases actively suppress multiple host cell activities that help the virus evade host defense mechanisms, promote viral replication and promote host cell apoptosis. For example, enterovirus proteases can cleave eukaryotic translation initiation factors 4GI (eIF4GI)5, 6, 7 and eIF4GII.8, 9 Picornavirus proteases have also been reported to cleave other canonical translation initiation factors in the cap-dependent translation initiation complex, such as eIF5B10 and eIF4A.11 Additionally, viral protease 3C has been demonstrated to cleave Ras-GAP SH3 domain-binding protein 1 (G3BP1), a key nucleating protein in stress granule formation, at late time points of infection.12, 13 G3BP1 cleavage causes stress granule disassembly12 and leads to the release of translation initiation factors that may be hijacked for viral polyprotein translation. Accumulating evidence implies that viral proteases have crucial roles in modulating viral and host gene expression through cleavage of various host protein factors involved in mRNA transcription and cap-dependent translation.
Death-associated protein 5 (DAP5) is a eukaryotic translation initiation factor that preferentially initiates cap-independent translation.14 This 97-kDa protein is homologous with the central region of canonical translation initiation factor eIF4GI. DAP5 contains binding sites for eIF4A and eIF3, but lacks an eIF4E site, the mRNA cap-binding protein.14 DAP5 initiates translation of proteins specifically expressed during development, cell cycle regulation and endoplasmic reticulum (ER) stress conditions, where global cap-dependent translation is compromised.15, 16 It has been reported that DAP5 regulates IRES-driven translation of Bcl-2 (B-cell lymphoma 2), p53, XIAP (X-linked inhibitor of apoptosis protein), CDK1 (cyclin-dependent kinase 1), c-Myc and other IRES-containing genes under conditions of stress, growth and apoptosis.17, 18, 19 DAP5 mRNA also contains an IRES in its 5′-UTR, and is proficient in promoting its own translation, generating a positive feedback loop for its expression.14, 15 Additionally, DAP5 may undergo post-translational modifications, such as cleavage by caspases during apoptosis, generating a truncated, yet functional 86-kDa form of DAP5 that is more potent and efficient at translation initiation.14
The role of DAP5 in initiation of IRES-driven translation has been studied in various model systems but not in the setting of viral infection. In the present study, we demonstrated that DAP5 is cleaved during CVB3 infection by protease 2A but not 3C. Further, we identified amino-acid reside G434 as the site of 2A cleavage. Once cleaved, DAP5 N-terminal truncate (DAP5-N) largely translocates into the nucleus, whereas the DAP5 C-terminal truncate (DAP5-C) remains primarily within the cytoplasm. Our data indicate that DAP5-N retains IRES-mediated translation; however, it differentially regulates the IRES-containing genes involved in cell growth and apoptosis compared with the wild type (WT) DAP5. Exogenous expression of DAP5-N or DAP5-C induces more cell death when compared with the WT DAP5. Further, viral capsid protein 1 (VP1) as well as viral titer is higher in DAP5-N-overexpressing cells during the late phase of infection (7 h post infection (hpi)) compared with that in WT (WT DAP5) or an uncleavable point mutant of DAP5 (glycine 434 to glutamic acid-mutated DAP5 (G434E DAP5)). Thus, viral protease 2A cleavage of DAP5 produces a cleavage product that robustly contributes to viral replication and viral-induced apoptosis, facilitating CVB3 progeny release.
Results
DAP5 is cleaved during CVB3 infection in tissue culture and in mouse heart
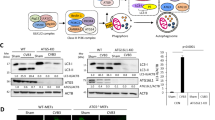
During CVB3 infection, we observed cleavage of DAP5 indicated by accumulation of ~45-kDa DAP5-N and ~52-kDa DAP5-C at 5 hpi in HeLa cells (Figures 1a and b). This finding suggests that DAP5 is not degraded during CVB3 infection, but cleaved into truncated fragments. However, at later time points, the sum of proform and cleavage products does not equal the total level of DAP5 early in infection, potentially because of altered expression of DAP5. To verify this, quantitative RT-PCR (q-RT-PCR) was performed. DAP5 transcription was downregulated at 5 and 6 hpi (Figure 1c). To investigate whether the cleavage of DAP5 also occurs in vivo, a CVB3-infected myocarditis mouse model was used. Mouse heart tissue was harvested at 3 and 9 days post infection (dpi). Cleavage of DAP5 to 45-kDa DAP5-N and 52-kDa DAP5-C fragments were observed (Figure 1d). In contrast to in vitro DAP5 expression, DAP5 was upregulated relative to controls in mouse hearts. This may be because of a lower percentage of CVB3-infected cells in the myocardium compared with that in tissue culture cells. This may also explain the relatively low abundance of DAP5 cleavage products in vivo versus in vitro, as viral infection is required for cleavage. Additionally, we found that at 9 dpi, heart tissue showed immune cell infiltration, that is, myocarditis (Figure 1e). Our findings indicate that the cleavage of DAP5 occurs both in vitro and in vivo.
DAP5 is cleaved into N- and C-terminal-truncated forms during CVB3 infection in vitro and in vivo and is transcriptionally downregulated. HeLa cells were infected with CVB3 at 10 MOI (multiplicity of infection) and collected at the indicated time points after infection. Lysates were analyzed by western blot with the indicated N-terminal (a) and C-terminal (b) DAP5 antibodies against the 45- and 52-kDa product, respectively. The line indicates ~50-kDa. *N- or C-terminal DAP5 cleavage product (cp). (c) DAP5 mRNA transcription levels were determined by q-RT-PCR using the total RNA isolated from cells described above. DAP5 mRNA is downregulated at 5 and 7 hpi in HeLa cells by ~40% relative to sham-infected controls. (d) A/J mice at 4 weeks of age were infected with CVB3 at 105 pfu or sham-infected with PBS as a control. Hearts were collected at 3 and 9 dpi. Harvested heart tissue was lysed and analyzed by western blot using the N- and C-terminal-specific antibody. GAPDH was used as a loading control. (e) Hematoxylin and eosin (H&E) staining of A/J mice heart tissue at 9 dpi. Protein levels of pro- and cleaved forms of DAP5 were quantitated by densitometry using the NIH ImageJ software (http://imagej.nih.gov/ij/index.html) and normalized to GAPDH or β-actin. Values are presented under each blot, with sham levels set to 1.00
Viral protease 2A but not 3C is responsible for DAP5 cleavage during CVB3 infection
To determine if viral proteases are responsible for cleavage of DAP5, plasmids expressing WT FLAG-tagged DAP5 and protease 2A or 3C via IRES-driven cap-independent translation were co-transfected into HeLa cells, and cleavage was determined by western blot. Our laboratory previously showed that caspases are unable to cleave DAP5 into 45- and 52-kDa fragments7 and that transient transfection of plasmids expressing CVB3 2A or 3C does not induce cleavage of DAP5. This was likely due to the use of a cap-dependent classical expression plasmid, which would be inactivated once the synthesized protease cleaved eIF4G and other canonical translation initiation factors. As 2A processivity of eIF4G may be higher compared with 2A processivity of DAP5, the loss of 2A expression may have mitigated any detectable result. To verify this, we co-transfected HeLa cells with WT DAP5 and pIRES-2A or pIRES-3C. The 45-kDa DAP5-N products were only observed in the 2A-expressing cells, suggesting that viral protease 2A is responsible for the cleavage of DAP5 during CVB3 infection (Figure 2a). To confirm that these plasmids expressed active 2A and 3C, the cleavage of endogenous, known substrates of 2A and 3C was measured. Figure 2b shows that endogenous DAP5 was cleaved by 2A but not 3C. Other known substrates, eIF5B and eIF4G,10, 20, 21 were cleaved either by 2A or 3C (Figures 2c and d). Additionally, the cleavage of eIF4G occurred at 24h, compared with 48 h after transfection for DAP5, suggesting that 2A processivity of eIF4G is in fact faster than 2A processivity of DAP5.
Viral protease 2A but not 3C cleaves overexpressed WT FLAG-DAP5 and endogenous DAP5 into 45- and 52-kDa truncates. (a) pIRES-2A or 3C plasmid was co-transfected into HeLa cells with WT FLAG-DAP5. As a control, HeLa cells were co-transfected with an empty pIRES vector and WT FLAG-DAP5 or with WT FLAG- DAP5 only. Cell lysates were collected 48 h after transfection and analyzed by western blot using an anti-FLAG antibody. β-actin was used as a loading control. (b) pIRES vector, pIRES-2A or pIRES 3C was transfected into HeLa cells. Lysates were collected for western blot analysis of endogenous 45-kDa DAP5-N. *DAP5 cleavage product (cp). Densitometric analysis was performed as indicated in Figure 1, with values presented under each blot. As a control, western blot was conducted on eIF4GI and eIF5B, to verify pIRES-2A and 3C constructs were effective at cleaving their respective substrates (c and d)
Glycine 434 is the site of DAP5 cleavage by 2A during CVB3 infection
According to the molecular mass of the cleavage products DAP5-N and DAP5-C, we proposed that the cleavage site is located upstream of the middle region of DAP5 protein (Figure 3a). Further, we narrowed down the potential DAP5 cleavage sites by using the reported 2A attack sites as a guide.22 Additionally, bioinformatics prediction using the NetPicoRNA 1.0 software (http://www.cbs.dtu.dk/services/NetPicoRNA/) (data not shown) was performed. Based on this available information, we proposed that glycine 434 might be the site for DAP5 cleavage. For verification, a point mutation changing G434E was constructed by site-directed mutagenesis (Figure 3b) in the FLAG-tagged DAP5 construct, referred to as G434E DAP5. Cells expressing WT DAP5 showed the 45-kDa cleavage product at the 6 hpi, whereas cells expressing G434E-DAP5 remained uncleaved (Figure 3c).
Glycine 434 is the site of 2A cleavage of DAP5 during CVB3 infection. (a) Schematic structures of eIF4G and DAP5 as well the known binding sites for proteins involved in translation initiation. The red dashed line indicates the proposed 2A cleave site at amino-acid residue 434 of DAP5 protein. (b) Sequence structure illustrating the site-directed mutagenesis at glycine 434 of DAP5. (c) HeLa cells were transfected with FLAG-tagged WT DAP5 construct or G434E DAP5 mutant for 48 h and subsequently infected with CVB3 at an MOI (multiplicity of infection) of 10 or sham-infected with PBS. Lysates were collected at 6 hpi and analyzed by western blot to detect DAP5 using an anti-FLAG antibody. *DAP5 cleavage product (cp). Densitometric analysis was performed as indicated in Figure 1, with values presented under each blot
DAP5-N translocates to the nucleus during ectopic 2A overexpression or CVB3 infection
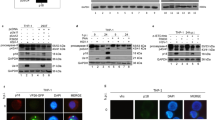
To determine if the subcellular localization of the DAP5 cleavage products differs from that of WT DAP5, FLAG-tagged DAP5-N (from N terminus to G434) and HA-tagged DAP5-C (from G434 to the C terminus) expression plasmids (Figure 4a) were generated. Following transfection, lysates were subjected to nuclear and cytoplasmic fractionation. WT DAP5 (100%) and DAP5-C (87%) remained mainly in the cytoplasm (Figure 4b). However, a significant portion of DAP5-N localized to the nucleus (45%). Further confirmation was provided by immunostaining. The ectopic expression of the cleavage products resulted in nuclear translocation only for DAP5-N (Figure 4c). Next, we determined whether ectopic expression of viral protease 2A could cause cleavage of DAP5 and subsequent nuclear translocation of the N-terminal portion of DAP5. Figure 4d demonstrates that in WT DAP5/pIRES-2A-transfected cells the N-terminal FLAG tag portion of WT DAP5 translocated to the nucleus, whereas the C-terminal HA portion remained in the cytoplasm. However, in G434E DAP5/pIRES-2A-transfected cells the majority of the DAP5 proteins were distributed in the cytoplasm, similar to cells expressing WT DAP5 co-transfected with empty vector. Last, WT DAP5-transfected cells show DAP5 nuclear translocation beginning at 5 hpi with native CVB3 (Figure 4e). These results demonstrate that the viral protease 2A cleavage of DAP5 at G434 during CVB3 infection results in a partial nuclear translocation of the N-terminal DAP5.
DAP5-N translocates to the nucleus, whereas G434E DAP5 and DAP5-C remain in the cytoplasm. (a) Schematic structures of clones of the DAP5-N and DAP5-C. The FLAG and HA tags as well as the binding sites of interacting proteins involved in translation initiation are indicated. The numbers are the specific amino-acid positions for the binding sites. (b) Western blot analysis of cellular distribution of WT DAP5, DAP5-N and DAP5-C after transfection. HeLa cells were transfected with WT DAP5 (FLAG), DAP5-N (FLAG) or DAP5-C (HA) for 48 h. Nuclear and cytoplasmic proteins were isolated using NE-PER Kit (Thermo Scientific). Histone-1 was used as a nuclear purity control and β-actin was used as a loading control. Densitometric analysis was performed as in Figure 1. Values are displayed as percentages of nuclear and cytoplasmic protein values. (c) Confocal imaging of intracellular localization of WT DAP5, DAP5-N and DAP5-C after transfection. HeLa cells were transfected as described in (b). Cells were immunostained with FLAG (WT and DAP5-N) or HA (DAP5-C) antibody and probed with Alexa Fluor 488 (green for WT and DAP5-C) or Alexa Fluor 594 (red for DAP5-N). DAPI staining was used to image nuclei (blue). (d) Confocal imaging of FLAG-WT DAP5-HA, or FLAG-G434E-DAP5, co-transfected with pIRES vector or protease 2A-expressing cells. HeLa cells were co-transfected with a plasmid expressing WT DAP5 tagged with N-terminal FLAG (columns 1 and 2) and a C-terminal HA (columns 5 and 6) and either pIRES empty vector or 2A-expressing pIRES-2A plasmid. Cells co-transfected with the G434E DAP5 and pIRES empty vector or pIRES-2A (columns 3 and 4) are additional controls. Cellular distribution of the cleavage products were detected by immunostaining for FLAG or HA using secondary goat anti-rabbit IgG labeled with Alexa Fluor 488 (green) and imaged by confocal microscopy. (e) Confocal imaging of intracellular localization of WT DAP5 (N-terminal FLAG, C-terminal HA-tagged) or G434E DAP5 in sham (columns 1 and 3) or CVB3-infected cells (columns 4 and 9). HeLa cells were transfected with plasmid for 48 h and subsequently infected with 10 MOI (multiplicity of infection) CVB3 for 5 or 7 h or sham-infected with PBS. Cellular distribution of WT DAP5-FLAG or -HA and FLAG-G434E DAP5 were detected by immunostaining of FLAG or HA and imaged by confocal microscopy as described for (d)
DAP5-N enhances VP1 production, while siRNA knockdown of DAP5 suppresses VP1 production
We sought to determine the effect of DAP5 cleavage on CVB3 replication. Cleavage percentages for each time point were calculated relative to proform and normalized to β-actin (Figure 5d). WT DAP5-transfected samples show DAP5 cleavage at 5 hpi (Figure 5a), whereas G434E DAP5-transfected samples show only full-length DAP5 with anti-FLAG antibody; anti-DAP5 does detect a small amount of cleaved endogenous DAP5 without FLAG tag (Figure 5a). Compared with empty vector-transfected cells, WT DAP5 induced 1.72-fold VP1 expression, whereas G434E DAP5 showed 0.48-fold expression (Figure 5e) at 7 hpi. This prompted us to examine the effect of DAP5-N and DAP5-C overexpression during CVB3 infection. Figure 5b demonstrates that ectopically expressed DAP5-N during CVB3 infection induces 2.5-fold VP1 translation compared with cells overexpressing DAP5-C, which remained equal to the vector (Figure 5f). These data suggest that viral protease 2A cleavage of DAP5 and subsequent generation of the N-terminal truncated form aid in viral replication. Data were further confirmed by knocking down DAP5-specific small interfering RNA (siDAP5) (Figure 5c), indicating that at 5 and 7 hpi, siRNA treatment induced 3.12- and 3.97-fold VP1 expression, respectively (Figure 5g).
Cleavage of DAP5 during CVB3 infection enhances VP1 production and siRNA knockdown of DAP5 suppresses VP1 production. (a) HeLa cells were transfected with pcDNA3.1 vector, WT DAP5 or G434E DAP5 for 48 h and subsequently infected with 10 MOI (multiplicity of infection) CVB3. Lysates were collected at the indicated time points and probed by western blot using antibodies against endogenous DAP5, FLAG-DAP5 and VP1. β-Actin was used as a loading control (b) HeLa cells were transfected with pcDNA3.1 vector, DAP5-N or DAP5-C and subsequently infected with 10 MOI CVB3. Lysates were collected at the indicated time points and probed by western blot using antibodies against FLAG (DAP5-N), HA (DAP5-C) or VP1. β-Actin was used as a loading control. (c) HeLa cells were transfected with DAP5-specific siRNA (siDAP5) to knockdown DAP5 expression and then infected with CVB3. Cell lysates were harvested at the indicated time points after infection for the analysis of DAP5 cleavage fragment and viral VP1 production. Scrambled small interfering RNA (scr) was used as a control. (d) DAP5 proform protein was quantified by densitometric analysis as described in Figure 1. Sham protein levels were set as 100% and subsequent values were calculated as percentages relative to sham protein levels. (e–g) VP1 protein production in (a–c) was quantified by densitometric analysis using the ImageJ (NIH) program and normalized to the corresponding controls and the data are presented as means±S.D. of three independent experiments in (d–f), respectively. *P<0.05 and **P<0.01
DAP5-N enhances viral particle formation, while siRNA knockdown of DAP5 decreases viral particle formation
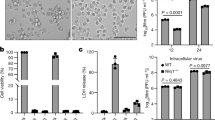
Given that WT DAP5 and DAP5-N enhance VP1 production compared with uncleavable G434E and DAP5-C, we sought to determine their effect on viral titer by viral plaque assay, using the supernatant of the cells described in Figure 5. WT DAP5-transfected cells show significantly more virus (data at 5 and 7 hpi: vector 5.33 × 105 and 2.74 × 106 plaque-forming units (pfu), WT 106 and 1.3 × 107 pfu, G434E 4.97 × 105 and 3.95 × 106 pfu; Figures 6a and b), indicating that cleavage of DAP5 enhances viral replication and progeny release. A similar increase was seen with DAP5-N (6.46 × 105 and 4.90 × 107 pfu). DAP5-C suppressed virus titer at 5 hpi (3.12 × 105 hpi) but not at 7 hpi (2.68 × 106 pfu; Figures 6c and d). Furthermore, siDAP5 significantly reduced viral particle formation (siDAP5 1.34 × 105 and 1.28 × 106 pfu at 5 and 7 hpi, scrambled 1.66 × 106 and 5.33 × 107 pfu; Figures 6e and f), indicating a significant role of DAP5 in viral particle formation and release during CVB3 infection.
Cleavage of DAP5 during CVB3 infection significantly enhances viral titer. (a and b) HeLa cells were transfected with WT DAP5, G434E DAP5 or empty vector for 48 h and subsequently infected with 10 MOI (multiplicity of infection) CVB3. Supernatants were collected at 5 (a) and 7 hpi (b) and used to measure CVB3 particles by plaque assay. (c and d) HeLa cells were transfected with DAP5-N, DAP5-C or empty vector for 48 h and subsequently infected with 10 MOI CVB3. Supernatants were collected at 5 (c) and 7 hpi (d) and used to measure CVB3 particles by plaque assay (e and f) HeLa cells were transfected with siDAP5 and then infected with 10 MOI CVB3. Scrambled small interfering RNA (scr) was used as a control. Supernatants were collected at 5 (e) and 7 hpi (f) and used to measure CVB3 particles by plaque assay. Data are presented as means±S.D. n=3, *P<0.05 or **P<0.01
DAP5-N and DAP5-C differentially regulate translation of p53 and Bcl-2 and result in apoptotic cell death
Previous studies have identified a role of DAP5 in the translation of IRES-containing genes Bcl-2 and p53.17, 18 Overexpression of WT-DAP5 and DAP5-N enhanced translation of p53 and Bcl-2, whereas DAP5-C increased p53 expression and decreased Bcl-2 expression (relative to control, p53: WT-DAP5 1.91, DAP5-N 1.73, DAP5-C 1.42; Bcl-2: WT-DAP5 2.33, DAP5-N 1.39, DAP5-C 0.54; Figures 7a and c). To study the mechanism(s) by which DAP5 and its cleavage products mediate cap-independent translation, phosphorylated (Ser209) eIF4E and total eIF4E were quantified (Figures 7a and d). To our surprise, overexpression of WT-DAP5, DAP5-N and DAP5-C significantly reduced eIF4E phosphorylation (7.94-, 8.68- and 10.27-fold relative to vector) compared with the total protein, indicating an inhibitory effect on cap-dependent translation. Differential alterations of p53 and Bcl-2 translation by DAP5 fragments contributed to the overall induction of apoptotic cell death as indicated by cleavage of procaspase-3, cleavage of poly-ADP-ribose polymerase (PARP; Figures 7b and e) and a significant reduction in cell survival (Figure 7f) when compared with vector or WT DAP5.
DAP5-N and DAP5-C differentially regulate translation of p53 and Bcl-2 and result in apoptotic cell death. HeLa cells were transfected with WT DAP5, DAP5-N, DAP5-C or vector. At 48 h after transfection, cells lysates were prepared for western blot analysis of proteins using the indicated antibodies (a and b). Quantification of western blot signals was conducted by densitometry analysis using the ImageJ program and normalized to β-actin, total eIF4E or proform of the proteins (c–e). (f) Cell viability was analyzed by MTS assay. Cell viability of WT DAP5-transfected cells was defined as 100% (control). Other data are presented as the percentage of the control. *P<0.05 and **P<0.01; n=3
DAP5-N and DAP5-C differentially alter translation but not transcription of IRES-containing genes p53 and Bcl-2
To directly demonstrate an effect of DAP5 cleavage products on IRES-driven translation of p53, Bcl-2 and CVB3 mRNA, bicistronic luciferase reporters were constructed by insertion of the 5′-UTR of these genes or the CVB3 genome between Renilla and firefly reporter genes (Figure 8a). WT DAP5 and DAP5-C enhanced translation of p53, Bcl-2, and CVB3 genome, whereas DAP5-N only significantly enhanced translation of p53 and CVB3 genome, not Bcl-2 (Figure 8b), consistent with data in Figure 7a. To rule out transcriptional changes, q-RT-PCR was performed to measure Bcl-2 and p53 using β-actin, a non-IRES-containing mRNA, as a baseline (Figure 8c). No significant differences in mRNA levels were observed among these groups, indicating that DAP5-N and DAP5-C do not directly affect transcription of p53 or Bcl-2 mRNA.
Overexpression of DAP5-N and DAP5-C alters translation but not transcription of IRES-containing genes p53 and Bcl-2. (a) Schematic structures of luciferase reporters. The 5′-UTR of Bcl-2, p53 or CVB3 genome was inserted into the site between Renilla and firefly luciferase coding regions on C49 plasmid. (b) HeLa cells were co-transfected with the luciferase reporter plasmid and a plasmid expressing WT DAP5, DAP5-N or DAP5-C. At 48 h after transfection, cell lysates were collected for luciferase assay to determine the relative luciferase activity. Results are shown as means±S.D. (N=15). *P<0.05 and **P<0.01. (c) q-RT-PCR to detect mRNA of p53 and Bcl-2. HeLa cells were transfected with empty vector or a plasmid expressing WT DAP5, DAP5-N or DAP5-C. At 48 h after transfection, total RNAs were isolated from the cells and used for q-RT-PCR to measure the mRNAs of IRES-containing genes, p53 and Bcl-2. β-Actin mRNA was used as a control for cap-containing gene. All values were normalized to GAPDH
Discussion
Enterovirus proteases, such as CVB3 2A, cleave translation initiation factors,5, 6, 7, 10, 20, 23, 24, 25, 26 leading to shutoff of cellular translation. DAP5, an eIF4G homolog responsible for IRES-mediated translation, was found to be cleaved by CVB3 protease 2A. The role of DAP5 in initiation of mRNA translation of IRES-containing genes in cellular stress conditions has been studied in different model systems.14, 15, 19, 27, 28 However, proteolytic modifications of DAP5 by viral proteases and subsequent functional changes have not been previously explored. By site-directed mutagenesis, FMKSQ↓G434LSQ was identified as the cleavage site of 2A protease, consistent with cleavage recognition sites reported for other substrates of 2A.22, 29 Intriguingly, cleavage of DAP5 caused partial nuclear translocation of the N-terminal cleavage product, whereas the C-terminal cleavage product remained primarily in the cytoplasm. Cleavage and the subsequent subcellular redistribution of the truncated proteins resulted in altered DAP5 function, analogous to that reported for caspase-3 cleavage of DAP5.14 Cleavage alters the cellular environment in a way that is advantageous for viral replication. This occurs in vitro and in vivo as demonstrated here. Knockdown of DAP5 during CVB3 infection resulted in significant reductions in VP1 levels and viral titer. Overexpression of DAP5-N resulted in higher levels of both VP1 and virus production. Upon cleavage, DAP5-N is still functional and has a role in differentially regulating translation. In addition, DAP5-N undergoes nuclear translocation upon cleavage during late CVB3 infection (a bioinformatically predicted nuclear localization signal is present), although the function following nuclear localization remains unknown. DAP5 reportedly translocates to the nucleus during all-trans retinoic acid (ATRA)-induced terminal differentiation in leukemia cells, resulting in the inhibition of phosphoinositide 3-kinase/protein kinase B (PI3K/Akt) prosurvival pathway.30 PI3K/Akt pathway suppression via dephosphorylation of Akt31 occurs concurrently with DAP5 nuclear translocation (6 hpi) during CVB3 infection, corresponding with the induction of apoptosis. This is similar to our finding showing induction of apoptosis, as evidenced by activation of procaspase-3 and reduction of cell survival following cleavage and nuclear translocation of DAP5-N. Future studies may identify the nuclear function of DAP5-N and explore the effect of DAP5 cleavage on immune cell infiltration and viral pathogenesis.
In this study, CVB3 2A protease cleavage of DAP5 produced two fragments of DAP5 protein: the 45-kDa DAP5-N remained active in IRES-driven translation initiation and favored the translation of proapoptotic genes (e.g. p53) and of CVB3, whereas the 52-kDa DAP5-C inhibited global translation and indirectly facilitated IRES-driven translation by dephosphorylation of eIF4E, the cap-binding translation initiation factor. Preferential enhancement by DAP5-N of p53 but not Bcl-2 may be attributed to DAP5-N-induced caspase activation, limiting the expression of Bcl-2 and thereby inducing apoptosis.32 DAP5-C induced nonspecific, cap-independent translation of Bcl-2, p53 and CVB3 likely through competition with eIF4E for MAP kinase signal-integrating kinase 1 (MNK1) phosphorylation, thus inhibiting eIF4E from participating in translation initiation.33, 34, 35 Changes in function may be explained based on the structural differences in each DAP5 fragment. DAP5 shares structural homology with the middle region of eIF4GI, termed the MIF4G domain. This 30-kDa MIF4G domain mediates protein–protein interactions with eIF4A and eIF3 and also exhibits RNA- and DNA-binding capabilities.36 It has been shown to interact directly with the IRES element of the encephalomyocarditis virus (EMCV) RNA37, 38 and allows eIF4G to recruit the ribosome to the EMCV RNA in a cap-independent manner by interacting with eIF3 and the RNA simultaneously. As DAP5-N retains the eIF4A- and eIF3-binding domains, it is competent for initiation of IRES-driven translation. This result is in line with previous reports that the C-terminal fragment of eIF4G drives IRES-containing mRNA translation.39, 40 This study showed that primary cleavage of eIF4G by picornavirus proteases generates an 18-kDa N-terminal polypeptide and a large C-terminal fragment. The C-terminal fragment possesses eIF4A- and eIF3-binding domains, is responsible for ribosome binding via eIF3 and, in complex with eIF4A, forms part of the RNA helicase apparatus.39, 40 This is further supported by another in vitro translation study using rabbit reticulocyte lysates,41 and recently, the interaction sites between DAP5, MIF4G and eIF4A have been identified by crystal structural analysis.42, 43 These data suggest that eIF4A- and eIF3-binding domains on DAP5-N are critical for cap-independent translation. DAP5-N-mediated translation of IRES-containing genes differs from WT DAP5. DAP5-N initiates translation of IRES-containing genes that are proapoptotic, as indicated by our results showing the enhancement of p53 IRES translation, cleavage of procaspase-3 and a significant reduction in cell survival rate. Furthermore, DAP5-C likely contributes to the enhancement of cell death by reduction of Bcl-2 and enhancement of IRES-mediated p53.
The function of DAP5-C may also relate to its structural features. It is documented that the C-terminal region of DAP5 shares structural homology with eIF4G. These homologous regions include MA3 (or mini-protein) domain and an MNK1-binding motif. However, the MA3 domain of DAP5 does not support eIF4A binding,44 leaving DAP5 with a single eIF4A interaction domain on the DAP5-N cleavage product, compared with two eIF4A-binding domains on protease 2A cleaved eIF4G.45 CVB3 and all type I viral IRES translation requires cleaved eIF4G (or DAP5) and eIF4A45 as eIF4G (and DAP5) bring eIF4A into the translation initiation complex. As such, eIF4G has a stoichiometric advantage (i.e. higher avidity) over DAP5.46 In combination with the higher affinity of 2A to eIF4G over DAP5,47 this may explain the relatively low changes in VP1 expression when DAP5-C is overexpressed. Although DAP5-C induces dephosphorylation of eIF4E, its effect on enhancement of viral translation may be modest, given that eIF4E is downregulated by microRNA 141 during enteroviral infection48 and eIF4E dephosphorylation occurs early during infection.34 However, the generation of DAP5-C, along with 4EBP1,49 may partially explain the dephosphorylation of eIF4E later during infection, where DAP5-C competes with eIF4E for MNK1 phosphorylation. In addition, the C-terminal regions of both DAP5 and eIF4G contain two aromatic and acidic boxes (AA boxes), also known as eIF5C or W2 domains.50, 51 Notably, both eIF4G and DAP5 interact with MNK1, which phosphorylates eIF4E, using the AA-box motif, when eIF4E is in the pretranslation initiation complex.35 In contrast, DAP5, but not eIF4G, binds to eIF2β through the AA box motif.52, 53 Therefore, DAP5-C is incapable of direct interaction with IRES mRNA owing to the lack of eIF4A-binding domain; however, it likely indirectly enhances IRES-driven translation by competing with eIF4E. Thus, DAP5-C may have a dominant-negative effect on cap-dependent translation and indirectly enhance cap-independent translation.
Taken together, this study has revealed for the first time that DAP5 is cleaved by protease 2A during infection, resulting in altered subcellular distribution and activity. The DAP5 cleavage products differentially regulate IRES-containing genes, promoting the translation of proapoptotic genes as well as viral genome, leading to enhanced viral replication and cell apoptosis.
Materials and Methods
Virus, cells, animals and plasmids
This study was carried out in strict accordance with the recommendations in the Guide to the Care and Use of Experimental Animals – Canadian Council on Animal Care. All protocols were approved by the Animal Care Committee, University of British Columbia (protocol number: A11-0052). CVB3 (CG) strain was obtained from Dr. Charles Gauntt (University of Texas Health Science Center, San Antonio, TX, USA) and propagated in HeLa cells (ATCC, Manassas, VA, USA). Virus stock was isolated from cells by three freeze–thaw cycles, followed by centrifugation to remove cell debris, and stored at −80oC. The titer of virus stock was determined by plaque assay as described in a later section. 4-Week old male A/J mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA). Mice were infected with 105 pfu of CVB3 or sham infected with PBS (Sigma, St. Louis, MO, USA) by intraperitoneal inoculation. HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Clontech, Palo Alto, CA, USA). FLAG-DAP5/97 plasmid (WT DAP5) was generously provided by Dr. Martin Holcik (Children’s Hospital of Eastern Ontario, Ottawa, ON, Canada).15 pIRES-2A was produced as described previously.12 Cell lystates were prepared at 24 and/or 48 h after transfection for western blot analysis using the appropriate antibody, described below. Two plasmids (pcDNA3-FLAG-DAP5N and pcDNA3-DAP5C-HA), expressing the N- and C-terminal cleavage products of DAP5, respectively, were constructed by a PCR-mediated method. Briefly, cDNA fragments encoding the N- or C-terminal region of DAP5 were synthesized by PCR using the up- and downstream primers containing XhoI/BamHI and XhoI/EcoRI restriction sites, respectively. The fragments were cloned into pcDNA3.1-5′-FLAG and pcDNA3.1-3′-HA vector at the above-mentioned restriction sites, respectively. The uncleavable DAP5 plasmid pcDNA3.1-FLAG-DAP5/G434E was constructed commercially (TopGene Technologies, Montreal, QC, Canada) by site-directed mutagenesis of the WT pcDNA3.1-FLAG-DAP5 plasmid to change nucleotides G to E, which resulted in the change from glycine at 434 to glutamic Acid. HeLa cells were transfected with WT DAP5 or G434E DAP5 and subsequently infected with CVB3 at 48 h after transfection. Cell lysates were analyzed by western blot using a FLAG antibody. The bicistronic luciferase reporter plasmids (C49-CVB3-5′-UTR, C49-Bcl2-5′-UTR, C49-p53-5′-UTR) were constructed by inserting the corresponding 5′-UTRs between the Renilla and the firefly luciferase coding regions on C49 vector, a kind gift from Dr. Joanna Floros’s laboratory (Hershey, PA, USA). The CVB3 and Bcl-2 5′-UTRs were amplified by PCR using specific primers and the plasmids we have as templates. The p53 5′-UTR18 was synthesized by RT-PCR and cloned into the C49 vector. All the 5′-UTRs are flanked by EcoRI digesting sites matching the cloning site on the vector.
Transfection and virus infection
HeLa cells (2 × 105/well) at 70–80% confluence in 6-well plates were washed with PBS and overlaid for 24 h with transfection complex containing 2 μg of plasmid DNA and 10 μl of Lipofectamine 2000 (Invitrogen, Waltham, MA, USA) or 10 μM scramble siRNA or DAP5 siRNA (Santa Cruz Biotechnology Inc., Dallas, TX, USA) and 5 μl Oligofectamine (Invitrogen) reagent per well. The transfection medium was then replaced with DMEM containing 10% FBS and the incubation was continued for 24 h before viral infection, or 48 h for plasmid DNA or 48 h for all siRNA experiments before harvesting. For viral infection, cells were washed with PBS and infected in 500 μl of serum-free DMEM at an MOI of 10 or sham-infected with PBS for the indicated time points in hours (hpi).
Western blot analysis
Western blots were performed using standard protocols as described previously.54 Mouse heart tissue was sectioned from the apical tissue and homogenized in MOSLB lysis buffer. The lysates were centrifuged at 13 000 × g for 20 min and the supernatants were collected for measuring the protein concentration using the Bradford Assay (Bio-Rad Laboratories, Mississauga, ON, Canada). Cells were washed two times in cold PBS after transfection or infection at different time points mentioned above. Cells were lysed in lysis buffer (0.025 M Tris-HCl, pH 8.0, 137 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100 and protease inhibitor cocktail) on ice for 20 min. Supernatants containing proteins were isolated by centrifugation at 13 000 × g at 4 °C for 15 min. Nuclear and cytoplasmic proteins were isolated using the NePER Kit according to the manufacturer’s instructions (Thermo Scientific, Waltham, MA, USA). WT DAP5, DAP5-N or DAP5-C was transfected into HeLa cells for 48 h in the absence of CVB3 infection, followed by nuclear protein extraction to separate the nuclear and cytoplasmic fractions. Histone-1 was used as a nuclear purity control and β-actin was used as a loading control, which is present in both the nucleus and cytoplasm.55, 56 Proteins were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes (Amersham GE, Buckinghamshire, UK). Membranes were blocked in 5% skim milk in Tris-buffered saline 10% Tween (TBST) buffer for 1 h and subsequently incubated with one of the following primary antibodies against DAP5-N (Thermo Scientific), DAP5-C (Cell Signaling Technologies, Danvers, MA, USA), Oct-8 (FLAG), HA and p53 (Santa Cruz Biotechnology Inc.), Bcl-2 (Cell Signaling Technologies), caspase-3 (Cell Signaling Technologies), PARP (Santa Cruz Biotechnology Inc.), phospho-eIF4E (Ser209) (Santa Cruz Biotechnology Inc.), total eIF4E (Santa Cruz Biotechnology Inc.), VP1 (Dako, Santa Clara, CA, USA) and β-actin (Sigma) at 4 °C overnight. Membranes were then washed in TBST three times for 10 min each, followed by incubation with the appropriate secondary antibody (goat anti-mouse or donkey anti-rabbit) conjugated to horseradish peroxidase (Amersham, Piscataway, NJ, USA) and visualized using the enhanced chemiluminescence method as per the manufacturer’s instructions (Amersham).
Quantitative RT-PCR
Cellular mRNAs were extracted and isolated using RNeasy Mini Kit (Qiagen, Venlo, Netherlands) according to the manufacturer’s instructions. Reverse transcription of RNAs was performed using SuperScript III First-Strand cDNA Synthesis System for RT-PCR according to the manufacturer’s instructions (Invitrogen). cDNA was measured by q-RT-PCR using the QuantiTect SYBR Green PCR Kit (Qiagen). Primers were designed from previous publications; for p53,18 Bcl-217 and DAP5,17 glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA levels served as an endogenous control. All real-time q-RT-PCR experiments were performed in triplicate with no-template as a negative control.
Bicistronic luciferase reporter assay
HeLa cells were seeded in 24-well plates and co-transfected with C49-CVB3-5′-UTR, C49-Bcl2-5′-UTR or C49-p53-5′-UTR and plasmid expressing pcDNA3.1 vector, WT DAP5, DAP5-N- or DAP5-C as described above. At 48 h after transfection, firefly and Renilla luciferase activities were measured using the Dual-Glo luciferase analysis system (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Briefly, the transfected cells were lysed with 100 μl/well of passive lysis buffer (Promega), and then 20 μl of the lysates were mixed with 100 μl of LAR II luciferase assay substrate. The firefly luciferase activity was measured immediately using a Tecan GENios fluorescence reader to detect the intensity of signal. One hundred microliters of Stop & Glo solution (Promega) was then added and the Renilla luciferase activity was detected using the same reader. All the assays were performed in triplicates.
Immunocytochemistry and confocal microscopy
HeLa cells proliferating on glass coverslips in a 6-well plate at ~70% confluence were co-transfected with pIRES-2A and plasmid (FLAG-DAP5-HA) expressing WT DAP5 containing an N-terminal FLAG tag and a C-terminal HA tag or G434E DAP5 (containing an N-terminal FLAG tag) and then immunostained the N- and C-terminal regions of DAP5 with FLAG and HA antibody, respectively, at 48 h after co-transfection or subjected to CVB3 infection at the indicated time points and subsequently immunostained as described previously.57 Briefly, cells were fixed with 4% paraformaldehyde, permeabilized in methanol/acetone (50:50) at −20 °C for 20 min and stained with an anti-FLAG or anti-HA primary antibody (Santa Cruz Biotechnology Inc.). Slides were washed and stained with a goat anti-rabbit IgG (H+L) labeled with Alexa Fluor 488 or 594 (Invitrogen). Nuclei were stained with 4′, 6′-diamidine-2′-phenylindole dihydrochloride (DAPI) (Vector Laboratories, Burlingame, CA, USA). Cells were observed with a Leica SP2 AOBS confocal microscope (Leica Microsystems, Wetzlar, Germany).
MTS assay
Cell viability was analyzed by using a 3-(4-5-dimethylthiazol-2-yl)-5-(-3-carboxymethoxyphenyl)-2H-tetrazolium salt (MTS) assay kit following the manufacturer’s instructions (Promega). Briefly, HeLa cells were transfected with a plasmid expressing WT, G434E, N-terminal or C-terminal DAP5 for 48 h. Cells were then incubated with MTS solution for 2 h. Absorbency of formazan was measured at 492 nm using enzyme-linked immunosorbent assay plate reader. The absorbency of vector-transfected cells was defined as 100% survival (control). The remaining data were converted to the percentage of control.
Viral plaque assay
Viral titers were determined as described previously.58 Briefly, HeLa cells were seeded into 6-well plates (8 × 105 cells per well) and incubated at 37 °C for 20 h to a confluence of ~0%, then washed with PBS and overlaid with 500 μl of virus serially diluted in cell culture medium. Virus was obtained by centrifugation (4000 × g) of freeze-thawed cell suspensions as described above. After a viral-adsorption period of 60 min at 37 °C, the supernatant was removed, the cells overlaid with 2 ml of sterilized soft Bacto-Agar minimal essential medium, cultured at 37 °C for 72 h, fixed with Carnoy’s fixative for 30 min and stained with 1% crystal violet. The plaques were counted and viral pfu/ml calculated.
Statistical analysis
Student’s t-test was used to analyze the data. Results are expressed as means±S.D of three independent experiments. A P-value <0.05 was considered statistically significant.
Abbreviations
- 5′-UTR:
-
5′-untranslated region
- AA boxes:
-
aromatic and acidic boxes
- Akt:
-
protein kinase B
- ATRA:
-
all-trans retinoic acid
- Bcl-2:
-
B-cell lymphoma 2
- CDK1:
-
cyclin-dependent kinase 1
- CVB3:
-
coxsackievirus B3
- dpi:
-
days post infection
- DAP5:
-
death-associated protein 5
- DAP5-C:
-
death-associated protein 5 C-terminal truncate
- DAP5-N:
-
death-associated protein 5 N-terminal truncate
- eIF:
-
eukaryotic translation initiation factor
- EMCV:
-
encephalomyocarditis virus
- ER:
-
endoplasmic reticulum
- G3BP1:
-
Ras-GAP SH3 domain-binding protein 1
- G434E DAP5:
-
glycine 434 to glutamic acid-mutated death-associated protein 5
- GAPDH:
-
glyceraldehyde 3-phosphate dehydrogenase
- hpi:
-
hours post infection
- IRES:
-
internal ribosome entry site
- MNK1:
-
MAP kinase signal-integrating kinase 1
- PARP:
-
poly-ADP-ribose polymerase
- PI3K:
-
phosphoinositide 3-kinase
- pfu:
-
plaque-forming units
- scr:
-
scrambled small interfering RNA
- siDAP5:
-
death-associated protein 5-specific small interfering RNA
- VP1:
-
viral capsid protein 1
- WT:
-
wild type
- XIAP:
-
X-linked inhibitor of apoptosis protein
References
Martino TA, Liu P, Sole MJ . Viral infection and the pathogenesis of dilated cardiomyopathy. Circ Res 1994; 74: 182–188.
Feldman AM, McNamara D . Myocarditis. N Engl J Med 2000; 343: 1388–1398.
Klump WM, Bergmann I, Muller BC, Ameis D, Kandolf R . Complete nucleotide sequence of infectious Coxsackievirus B3 cDNA: two initial 5' uridine residues are regained during plus-strand RNA synthesis. J Virol 1990; 64: 1573–1583.
Yang D, Wilson JE, Anderson DR, Bohunek L, Cordeiro C, Kandolf R et al. In vitro mutational and inhibitory analysis of the cis-acting translational elements within the 5' untranslated region of coxsackievirus B3: potential targets for antiviral action of antisense oligomers. Virology 1997; 228: 63–73.
Lamphear BJ, Kirchweger R, Skern T, Rhoads RE . Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem 1995; 270: 21975–21983.
Lamphear BJ, Rhoads RE . A single amino acid change in protein synthesis initiation factor 4G renders cap-dependent translation resistant to picornaviral 2A proteases. Biochemistry 1996; 35: 15726–15733.
Chau DH, Yuan J, Zhang H, Cheung P, Lim T, Liu Z et al. Coxsackievirus B3 proteases 2A and 3C induce apoptotic cell death through mitochondrial injury and cleavage of eIF4GI but not DAP5/p97/NAT1. Apoptosis 2007; 12: 513–524.
Svitkin YV, Gradi A, Imataka H, Morino S, Sonenberg N . Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J Virol 1999; 73: 3467–3472.
Gradi A, Svitkin YV, Imataka H, Sonenberg N . Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA 1998; 95: 11089–11094.
de Breyne S, Bonderoff JM, Chumakov KM, Lloyd RE, Hellen CU . Cleavage of eukaryotic initiation factor eIF5B by enterovirus 3C proteases. Virology 2008; 378: 118–122.
Belsham GJ, McInerney GM, Ross-Smith N . Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J Virol 2000; 74: 272–280.
Fung G, Ng CS, Zhang J, Shi J, Wong J, Piesik P et al. Production of a dominant-negative fragment due to G3BP1 cleavage contributes to the disruption of mitochondria-associated protective stress granules during CVB3 infection. PLoS One 2013; 8: e79546.
Fung G, Shi J, Deng H, Hou J, Wang C, Hong A et al. Cytoplasmic translocation, aggregation, and cleavage of TDP-43 by enteroviral proteases modulate viral pathogenesis. Cell Death Differ 2015; 58: 1–11.
Henis-Korenblit S, Strumpf NL, Goldstaub D, Kimchi A . A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol Cell Biol 2000; 20: 496–506.
Lewis SM, Cerquozzi S, Graber TE, Ungureanu NH, Andrews M, Holcik M . The eIF4G homolog DAP5/p97 supports the translation of select mRNAs during endoplasmic reticulum stress. Nucleic Acids Research 2008; 36: 168–178.
Yamanaka S, Zhang XY, Maeda M, Miura K, Wang S, Farese RV Jr. et al. Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J 2000; 19: 5533–5541.
Marash L, Liberman N, Henis-Korenblit S, Sivan G, Reem E, Elroy-Stein O et al. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol Cell 2008; 30: 447–459.
Weingarten-Gabbay S, Khan D, Liberman N, Yoffe Y, Bialik S, Das S et al. The translation initiation factor DAP5 promotes IRES-driven translation of p53 mRNA. Oncogene 2014; 33: 611–618.
Henis-Korenblit S, Shani G, Sines T, Marash L, Shohat G, Kimchi A . The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc Natl Acad Sci USA 2002; 99: 5400–5405.
Joachims M, Van Breugel PC, Lloyd RE . Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol 1999; 73: 718–727.
Carthy CM, Granville DJ, Watson KA, Anderson DR, Wilson JE, Yang D et al. Caspase activation and specific cleavage of substrates after coxsackievirus B3-induced cytopathic effect in HeLa cells. J Virol 1998; 72: 7669–7675.
Wong J, Zhang J, Yanagawa B, Luo Z, Yang X, Chang J et al. Cleavage of serum response factor mediated by enteroviral protease 2A contributes to impaired cardiac function. Cell Res 2012; 22: 360–371.
Kerekatte V, Keiper BD, Badorff C, Cai A, Knowlton KU, Rhoads RE . Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J Virol 1999; 73: 709–717.
Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N . Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 2005; 19: 104–113.
Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE . Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol Cell Biol 2004; 24: 1779–1790.
Cui W, Tao J, Wang Z, Ren M, Zhang Y, Sun Y et al. Neuregulin1beta1 antagonizes apoptosis via ErbB4-dependent activation of PI3-kinase/Akt in APP/PS1 transgenic mice. Neurochem Res 2013; 38: 2237–2246.
Warnakulasuriyarachchi D, Cerquozzi S, Cheung HH, Holcik M . Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J Biol Chem 2004; 279: 17148–17157.
Nevins TA, Harder ZM, Korneluk RG, Holcik M . Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J Biol Chem 2003; 278: 3572–3579.
Blom N, Hansen J, Blaas D, Brunak S . Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci 1996; 5: 2203–2216.
Ozpolat B, Akar U, Zorrilla-Calancha I, Vivas-Mejia P, Acevedo-Alvarez M, Lopez-Berestein G . Death-associated protein 5 (DAP5/p97/NAT1) contributes to retinoic acid-induced granulocytic differentiation and arsenic trioxide-induced apoptosis in acute promyelocytic leukemia. Apoptosis 2008; 13: 915–928.
Zhang HM, Qiu Y, Ye X, Hemida MG, Hanson P, Yang D . P58(IPK) inhibits coxsackievirus-induced apoptosis via the PI3K/Akt pathway requiring activation of ATF6a and subsequent upregulation of mitofusin 2. Cell Microbiol 2014; 16: 411–424.
Hanson PJ, Zhang HM, Hemida MG, Ye X, Qiu Y, Yang D . IRES-dependent translational control during virus-induced endoplasmic reticulum stress and apoptosis. Front Microbiol 2012; 3: 92.
Montero H, Garcia-Roman R, Mora SI . eIF4E as a control target for viruses. Viruses 2015; 7: 739–750.
Kleijn M, Vrins CL, Voorma HO, Thomas AA . Phosphorylation state of the cap-binding protein eIF4E during viral infection. Virology 1996; 217: 486–494.
Walsh D, Mathews MB, Mohr I . Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harbor Perspect Biol 2013; 5: a012351.
Ponting CP . Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem Sci 2000; 25: 423–426.
Pestova TV, Shatsky IN, Hellen CU . Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol 1996; 16: 6870–6878.
Lomakin IB, Hellen CU, Pestova TV . Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol Cell Biol 2000; 20: 6019–6029.
Rogers Jr GW, Richter NJ, Lima WF, Merrick WC . Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem 2001; 276: 30914–30922.
Oberer M, Marintchev A, Wagner G . Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev 2005; 19: 2212–2223.
Ohlmann T, Rau M, Pain VM, Morley SJ . The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J 1996; 15: 1371–1382.
Frank F, Virgili G, Sonenberg N, Nagar B . Crystallization and preliminary X-ray diffraction analysis of the MIF4G domain of DAP5. Acta Crystallogr Sec F 2010; 66 (Pt 1): 15–19.
Virgili G, Frank F, Feoktistova K, Sawicki M, Sonenberg N, Fraser CS et al. Structural analysis of the DAP5 MIF4G domain and its interaction with eIF4A. Structure 2013; 21: 517–527.
Imataka H, Sonenberg N . Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol 1997; 17: 6940–6947.
de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU . Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci USA 2009; 106: 9197–9202.
Padrick SB, Deka RK, Chuang JL, Wynn RM, Chuang DT, Norgard MV et al. Determination of protein complex stoichiometry through multisignal sedimentation velocity experiments. Anal Biochem 2010; 407: 89–103.
Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW . Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem 1982; 257: 14806–14810.
Ho BC, Yu SL, Chen JJ, Chang SY, Yan BS, Hong QS et al. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 2011; 9: 58–69.
Li X, Li Z, Zhou W, Xing X, Huang L, Tian L et al. Overexpression of 4EBP1, p70S6K, Akt1 or Akt2 differentially promotes Coxsackievirus B3-induced apoptosis in HeLa cells. Cell Death Dis 2013; 4: e803–e809.
Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG . Conserved bipartite motifs in yeast eIF5 and eIF2Bepsilon, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J 1999; 18: 1673–1688.
Bellsolell L, Cho-Park PF, Poulin F, Sonenberg N, Burley SK . Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1. Structure 2006; 14: 913–923.
Lee SH, McCormick F . p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. EMBO J 2006; 25: 4008–4019.
Liberman N, Gandin V, Svitkin YV, David M, Virgili G, Jaramillo M et al. DAP5 associates with eIF2beta and eIF4AI to promote internal ribosome entry site driven translation. Nucleic Acids Res 2015; 43: 3764–3775.
Liu Z, Zhang HM, Yuan J, Lim T, Sall A, Taylor GA et al. Focal adhesion kinase mediates the interferon-gamma-inducible GTPase-induced phosphatidylinositol 3-kinase/Akt survival pathway and further initiates a positive feedback loop of NF-kappaB activation. Cell Microbiol 2008; 10: 1787–1800.
Hofmann WA, de Lanerolle P . Nuclear actin: to polymerize or not to polymerize. J Cell Biol 2006; 172: 495–496.
McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ . Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol 2006; 172: 541–552.
Sall A, Zhang HM, Qiu D, Liu Z, Yuan J, Liu Z et al. Pro-apoptotic activity of mBNIP-21 depends on its BNIP-2 and Cdc42GAP homology (BCH) domain and is enhanced by coxsackievirus B3 infection. Cell Microbiol 2010; 12: 599–614.
Yuan J, Liu Z, Lim T, Zhang H, He J, Walker E et al. CXCL10 inhibits viral replication through recruitment of natural killer cells in coxsackievirus B3-induced myocarditis. Circ Res 2009; 104: 628–638.
Acknowledgements
We thank Dr. Adi Kimchi (Weizmann Institute of Science, Rehovot, Israel) and Dr. Martin Holcik (Apoptosis Research Centre, Children’s Hospital of Eastern Ontario) for providing us the plasmid Bcl-2 luciferase reporter and WT-FLAG-DAP5, respectively. We also thank Angela Chang and Erika Jang for their technical help (University of British Columbia). This work was supported by a grant-in-aid from the Canadian Institutes of Health Research (MOP-125995). Dr. Maged Hemida is a recipient of the CIHR-IMPACT and Heart and Stroke Foundation of Canada postdoctoral fellowship. XY is supported by a UGF Award from the University of British Columbia.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Willis
Rights and permissions
About this article
Cite this article
Hanson, P., Ye, X., Qiu, Y. et al. Cleavage of DAP5 by coxsackievirus B3 2A protease facilitates viral replication and enhances apoptosis by altering translation of IRES-containing genes. Cell Death Differ 23, 828–840 (2016). https://doi.org/10.1038/cdd.2015.145
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2015.145