Abstract
6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) is a master regulator of glycolysis by its ability to synthesize fructose-2,6-bisphosphate, a potent allosteric activator of 6-phosphofructo-1-kinase. Being a substrate of the E3 ubiquitin ligase anaphase-promoting complex-Cdh1 (APCCdh1), PFKFB3 is targeted to proteasomal degradation in neurons. Here, we show that activation of N-methyl-D-aspartate subtype of glutamate receptors (NMDAR) stabilized PFKFB3 protein in cortical neurons. Expressed PFKFB3 was found to be mainly localized in the nucleus, where it is subjected to degradation; however, expression of PFKFB3 lacking the APCCdh1-targeting KEN motif, or following NMDAR stimulation, promoted accumulation of PFKFB3 and its release from the nucleus to the cytosol through an excess Cdh1-inhibitable process. NMDAR-mediated increase in PFKFB3 yielded neurons having a higher glycolysis and lower pentose-phosphate pathway (PPP); this led to oxidative stress and apoptotic neuronal death that was counteracted by overexpressing glucose-6-phosphate dehydrogenase, the rate-limiting enzyme of the PPP. Furthermore, expression of the mutant form of PFKFB3 lacking the KEN motif was sufficient to trigger oxidative stress and apoptotic death of neurons. These results reveal that, by inhibition of APCCdh1, glutamate receptors activation stabilizes PFKFB3 thus switching neuronal metabolism leading to oxidative damage and neurodegeneration.
Similar content being viewed by others
Main
In contrast to the neuroprotective actions of mild glutamatergic synaptic activity,1 persistent activation of the N-methyl-D-aspartate subtype of glutamate receptors (NMDAR) – including the extra-synaptic ones2 – is known to underlie the pathogenesis of a number of neurological disorders, including Alzheimer's disease, amyotrophic lateral sclerosis or stroke.3 This phenomenon, known as excitotoxicity, causes neuronal apoptotic death through a not yet fully understood mechanism, but it is thought to involve an increase in intracellular Ca2+ through NMDAR, followed by plasma membrane depolarization and, hence, opening of voltage-gated Ca2+ channels, release from intracellular stores and reversal of the plasma membrane Na+/Ca2+ exchanger.4 This process eventually triggers the accumulation of mitochondrial Ca2+, leading to increased reactive oxygen species (ROS) formation, mitochondrial energy dysfunction, permeability transition pore opening and cytochrome c release.5 Besides mitochondria, it has also been recently shown that cytoplasmic NADPH oxidase has a key role in ROS production upon NMDAR stimulation.6 Thus, Ca2+ influx activates protein kinase C, which in turn phosphorylates and activates p47phox; p47phox coordinates NAPDH oxidase subunit organization, leading to enzyme activation.6 Regardless the origin of ROS, it is thought that neurons are highly vulnerable to mitochondrial stress, likely because of their inability to sufficiently activate glycolysis and, hence, to transiently compensate energy deficiency.7, 8 In contrast to neurons, astrocytes and other proliferative cells readily invoke glycolysis as a cytoprotective mechanism.8, 9, 10, 11
Glycolysis is controlled by the activity of 6-phosphofructo-1-kinase, the activity of which is highly dependent on its potent allosteric activator, fructose-2,6-bisphosphate (F2,6P2); in the brain, F2,6P2 biosynthesis almost exclusively relies on 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) activity.12, 13 Previously, we reported that the inability of neurons to promote cytoprotective glycolysis is because of the virtual absence of PFKFB3.14 Furthermore, we recently found that PFKFB3, through its KEN motif, is a substrate of the E3 ubiquitin ligase, anaphase-promoting complex (APC)-Cdh1 (APCCdh1),15 which accounts for the high instability of PFKFB3 and low glycolytic rate in neurons.15
Inhibition of APCCdh1 in postmitotic neurons triggers an accumulation of its substrate, cyclin B1, which mediates apoptotic death.16 Moreover, cyclin B1 accumulation can also be recapitulated by NMDAR stimulation, which activates cyclin-dependent kinase 5 (Cdk5)-mediated Cdh1 phosphorylation, leading to APCCdh1 inhibition.17 In view of the control that APCCdh1 exerts over PFKFB3 stability,15 here we hypothesized whether NMDAR stimulation, via APCCdh1 inhibition,17 regulates PFKFB3 protein levels in neurons. We show that NMDAR activation, through inhibition of APCCdh1 caused PFKFB3 stabilization leading to increased glycolysis and reduced activity of the pentose-phosphate pathway (PPP). This metabolic alteration triggered oxidative damage and excitotoxic neuronal death, thus suggesting that modulators of neuronal energy metabolism should be considered as targets in therapeutic strategies against neurodegenerative diseases.
Results
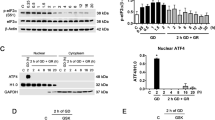
In order to test whether rat primary cortical neurons in culture responded to glutamate receptor activation, we first monitorized the changes in Fura-2 fluorescence. As shown in Figure 1a, Fura-2 F335/F363 ratio – an index of intracellular Ca2+ levels – increased by ∼1.3-fold immediately after the addition of glutamate (100 μM) or N-methyl-D-aspartate (NMDA; 100 μM). Furthermore, pre-incubation of neurons with the NMDAR antagonist, MK801 (10 μM), prevented by ∼60% glutamate-mediated increase in F335/F363 ratio (Figure 1a, left panel); MK801 almost abolished (by ∼90%) NMDA-mediated increase in F335/F363 ratio (Figure 1a, right panel). These results suggest that, under our experimental conditions – cells grown in serum-based medium – cortical neurons express functional NMDAR and, hence, are useful for the study of excitotoxic-mediated metabolic changes. To investigate whether glutamate receptor activation controls PFKFB3 stability, we then incubated neurons with glutamate (100 μM/15 min), and the levels of PFKFB3 protein were analyzed by western blotting. As shown in Figure 1b, glutamate triggered a time-dependent increase in PFKFB3, an effect that was maximal (by ∼2.1-fold) after 6 h. To see whether this effect was mediated by NMDAR, neurons were incubated with NMDA (100 μM/15 min), and PFKFB3 protein levels analyzed 6 h later. As depicted in Figure 1c, NMDA mimicked glutamate at increasing PFKFB3. Moreover, incubation of neurons with MK801 prevented glutamate-mediated increase in PFKFB3 (Figure 1d). Glutamate did not alter PFKFB3 mRNA levels (Figure 1e). These results suggest that activation of NMDAR stabilizes PFKFB3 protein in neurons.
Activation of NMDAR stabilizes PFKFB3 protein in neurons. (a) Incubation of rat primary cortical neurons with glutamate (left panel) or NMDA (right panel) increased the ratio of Fura-2-dependent fluorescence (at 510 nm) obtained after excitation at 335/363 nm (F335/F363), indicating an increase in intracellular Ca2+. MK801 (10 μM) partially prevented glutamate-induced changes in F335/F363 ratio and most of NMDA-dependent F335/F363 ratio changes. (b) Incubation of neurons with glutamate (100 μM/15 min) triggered time-dependent increase in PFKFB3 protein, which was maximal after 6 h. (c) NMDA (100 μM/15 min) mimicked glutamate at increasing PFKFB3. (d) NMDA receptor antagonist, MK801 (10 μM), prevented glutamate-mediated increase in PFKFB3. (e) Glutamate (100 μM/15 min) did not change PFKFB3 mRNA levels, as revealed by the unaltered intensity of the predicted 300 bp band after reverse-transcription of total RNA samples, followed by polymerase chain reaction (RT-PCR) using specific oligonucleotides for PFKFB3; GAPDH (279 bp band) was used as loading control; the black/white inverted images of the agarose gels are shown; w/o RT, RT-PCR for PFKFB3 without reverse transcriptase
Next, we investigated the involvement of APCCdh1 activity in determining PFKFB3 stabilization by NMDAR. In view that NMDAR activation promotes APCCdh1 inhibition by Cdk5-mediated phosphorylation of Cdh1,17 we tested whether this observation could be confirmed in our conditions. As shown in Figure 2a, glutamate (100 μM/15 min) promoted, after 4 h, H1 phosphorylation in neuronal samples immunoprecipitated with anti-Cdk5; furthermore, this effect was prevented by MK801, suggesting NMDAR-mediated activation of Cdk5 activity. In addition, Cdh1 was phosphorylated – suggesting APCCdh1 inhibition – 6 h after glutamate treatment, an effect that was also prevented by MK801 (Figure 2b). To further investigate if APCCdh1 activity regulated PFKFB3 stability upon glutamate receptor stimulation, neurons were transfected with a green fluorescent protein (GFP)-PFKFB3 construct to visualize PFKFB3 subcellular localization by confocal microscopy. PFKFB3 was mainly localized in the nucleus of neurons, but glutamate treatment promoted its accumulation, as revealed by the spread (nuclear plus cytosolic) localization (Figures 2c and d). Interestingly, Cdh1 overexpression prevented this effect, suggesting that deficiency in active Cdh1 was responsible for glutamate-mediated PFKFB3 spreading (Figures 2c and d). Furthermore, expression of a GFP-PFKFB3 form with its KEN box mutated to AAA (mut-PFKFB3), hence, insensitive to APCCdh1 activity,15 showed the spread-like localization, regardless of glutamate treatment (Figures 2c and d). Thus, glutamate-mediated PFKFB3 stabilization occurs via APCCdh1 inhibition.
Glutamate-mediated PFKFB3 stabilization occurs via Cdk5-mediated inhibition of APCCdh1 activity. (a) Glutamate treatment (100 μM/15 min) increased, after 1 h, Cdk5-mediated H1 phosphorylation in rat primary cortical neurons; this effect was fully abolished by MK801 (10 μM). (b) Cdh1 is phosphorylated 6 h after glutamate treatment (100 μM/15 min), an effect that was prevented by MK801 (10 μM). (c) Confocal microscopy images of neurons transfected with GFP-PFKFB3 reveals its nuclear localization. Glutamate promotes PFKFB3 accumulation, as revealed by its spread (nuclear plus cytosolic) localization; Cdh1 overexpression prevented this effect. GFP-PFKFB3 mutated on its KEN box (KEN → AAA; mut-PFKFB3) showed the spread-like localization, regardless of glutamate treatment. (d) Percentage of neurons showing nuclear or spread GFP-PFKFB3 localization in the experiments shown in c; these data were obtained by analyzing ∼30 neurons per condition per neuronal preparation (n=4). *P<0.05 versus the corresponding (nuclear or cytoplasmic) PFKFB3-none condition (ANOVA)
To elucidate whether NMDAR-mediated PFKFB3 protein stabilization had functional consequences for neuronal metabolism, we assessed the rates of glycolysis and PPP, as well as the glutathione redox status. The efficacy of a small interfering RNA against PFKFB3 (siPFKFB3) to prevent PFKFB3 protein accumulation was first tested. To do so, primary neurons were transfected with the GFP-PFKFB3 complementary DNA (cDNA) construct, and PFKFB3 protein was determined using an anti-flag (anti-GFP) antibody. As shown in Figure 3a, PFKFB3 was accumulated 6 h after glutamate (100 μM/15 min) incubation in control neurons (siControl); however, transfection of neurons with an siPFKFB3 decreased PFKFB3 protein abundance in control neurons and prevented glutamate-mediated PFKFB3 accumulation. We then assessed the rate of glycolysis, and we found it to be significantly enhanced, after 6 h, by glutamate treatment (Figure 3b); moreover, this was abolished by preventing PFKFB3 accumulation using the siPFKFB3 (Figure 3b). In view that glycolysis and PPP are two interconnected metabolic pathways, we then assessed whether the increase in glycolysis altered glucose utilization through the PPP. We found that glutamate treatment decreased, after 6 h, the rate of PPP, an effect that was abolished by siPFKFB3 (Figure 3c). Thus, glutamate triggers a PFKFB3-dependent increase in glycolysis and decrease in the PPP.
Glutamate stimulates PFKFB3-dependent increase in glycolysis, a decrease in PPP and promotes glutathione oxidation in neurons. (a) Incubation of GFP-PFKFB3-expressing neurons with glutamate (100 μM/15 min) induced, 6 h after treatment, PFKFB3 accumulation in control, siRNA-treated neurons (siControl), as revealed by an anti-GFP (Flag) antibody; transfection of neurons with an siPFKFB3 efficiently reduced PFKFB3 protein and prevented glutamate-induced PFKFB3 accumulation. (b) Incubation of neurons with glutamate (100 μM/15 min) increased, after 6 h, the rate of glycolysis, as assessed by the determination of [3-3H]glucose incorporation into 3H2O; this effect was abolished by preventing PFKFB3 accumulation in neurons previously transfected with siPFKFB3. (c) Glutamate treatment decreased, after 6 h, the rate of the PPP, as assessed by the determination of the difference between 14CO2 produced by [1-14C]glucose and that of [6-14C]glucose; this effect was abolished by siPFKFB3. (d) Glutamate treatment did not change GSx (left panel), but it increased GSSG (middle panel) and the oxidized glutathione redox status (GSSG/GSx; right panel); these effects were partially prevented by siPFKFB3. *P<0.05 versus none; #P<0.05 versus the corresponding siControl (ANOVA)
It has been previously shown that glucose metabolism through the PPP is neuroprotective15, 18, 19 because of its NADPH-regenerating function. Thus, NADPH is an essential cofactor for glutathione regeneration, hence, the PPP becomes necessary to prevent neuronal death by oxidative stress.18 Thus, we next aimed to elucidate whether the metabolic PPP/glycolytic shift triggered by glutamate treatment induced oxidative stress. As shown in Figure 3d, total glutathione (GSx) was unaltered, but its oxidized form (GSSG) and the glutathione oxidized status (GSSG/GSx ratio) significantly increased 6 h after glutamate treatment, and these effects were prevented by siPFKFB3. To further support evidence for oxidative stress, we next evaluated whether a putative increased ROS production by glutamate could be rescued by either knocking down a key glycolytic enzyme, phosphoglucose isomerase (PGI), or by overexpressing glucose-6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme of the PPP that we have previously shown to be efficient in neurons.15, 18 The efficacy of these tools were first tested by western blotting (Figure 4a); thus, transfection of neurons with the siRNA against PGI efficiently knocked down PGI protein, whereas overexpression of the cDNA encoding G6PD increased neuronal G6PD abundance. Glutamate treatment (100 μM/15 min) increased, after 6 h, neuronal ROS, an effect that was prevented by knocking down PGI or PFKFB3, as it was by overexpressing G6PD, or by blocking NMDAR with MK801 (Figure 4b). To assess neuronal vulnerability to oxidative stress in this paradigm, we then analyzed the proportion of annexin V+/7AAD− neurons (indicating neurons that had been targeted to apoptosis) after glutamate treatment. We found that glutamate increased, though modestly, apoptotic neuronal death via a mechanism that could be prevented by silencing PGI or PFKFB3, as well as by overexpressing G6PD or blocking NMDAR (Figure 4c). Together, these results indicate that NMDAR activation triggers oxidative stress and targets neurons for apoptotic death by shifting PPP to glycolysis.
NMDAR activation triggers oxidative stress and apoptotic death by switching PPP to glycolysis. (a) Transfection of neurons with an siRNA against PGI (siPGI), efficiently knocked down PGI protein abundance (left panel). Transfection of neurons with the full-length DNA encoding G6PD efficiently increased G6PD protein abundance (right panel). (b) Glutamate treatment (100 μM/15 min) increased ROS in neurons, as assessed by MitoSox fluorescence by flow cytometry; this effect was prevented by knocking down PGI (siPGI) or PFKFB3 (siPFKFB3), overexpressing G6PD, or blocking NMDAR with MK801 (10 μM). (c) Glutamate treatment increased apoptotic neuronal death, as assessed by annexin V+/7-AAD− fluorescence by flow cytometry; this effect was prevented by silencing PGI (siPGI) or PFKFB3 (siPFKFB3), overexpressing G6PD or blocking NMDAR with MK801. *P<0.05 versus none; #P<0.05 versus siControl (glutamate; 5 × 104 events were acquired in triplicate; results mean±S.E.M. from three independent neuronal preparations, n=3; ANOVA)
Finally, we sought to elucidate whether APCCdh1 activity was responsible for PFKFB3-mediated oxidative stress and neurodegeneration in excitotoxicity. As shown in Figure 5a, glutamate increased ROS in neurons transfected with low levels of wild-type PFKFB3 cDNA. However, transfection of neurons with identical cDNA amounts of the KEN box-mutant form of PFKFB3 (mut-PFKFB3) increased ROS to levels similar to those triggered by glutamate; moreover, glutamate did not further enhance ROS in neurons expressing mut-PFKFB3 (Figure 5a). Interestingly, apoptotic death triggered by glutamate in neurons transfected with PFKFB3 was mimicked by mut-PFKFB3 (Figure 5b). Thus, expression of APCCdh1-insensitive PFKFB3 mimics glutamate at causing oxidative stress and neuronal death.
Expression of APCCdh1-insensitive PFKFB3 mimics glutamate at causing oxidative stress and neuronal death. (a) Glutamate treatment (100 μM/15 min) increased ROS in neurons transfected with low levels of wild-type PFKFB3 cDNA; transfection of neurons with identical cDNA amounts of the KEN box-mut-PFKFB3 increased ROS to similar levels to those triggered by glutamate; glutamate did not further enhance ROS in neurons expressing mut-PFKFB3. (b) Glutamate increased apoptotic death of neurons transfected with low levels of PFKFB3 cDNA; transfection of neurons with identical cDNA amounts of mut-PFKFB3 increased apoptotic death to similar levels to those triggered by glutamate; glutamate did not further enhance apoptotic death in neurons expressing mut-PFKFB3. *P<0.05 versus none (PFKFB3; 3 × 104 events were acquired in triplicate; results mean±S.E.M. from three independent neuronal preparations, n=3; ANOVA)
Discussion
Neurons continuously degrade the glycolytic-promoting enzyme PFKFB3 by APCCdh1 activity, and this allows a considerable proportion of glucose to be oxidized via the PPP, which functions as an antioxidant and survival metabolic pathway.15 Here, we show that a short-term activation of glutamate receptors in cortical neurons triggers delayed, time-dependent PFKFB3 protein accumulation; the lack of change in the PFKFB3 mRNA abundance ruled out a transcriptional effect. Moreover, NMDA mimicked glutamate, and NMDAR antagonist MK801 prevented PFKFB3 accumulation, indicating the direct involvement of NMDAR stimulation in PFKFB3 stabilization. Previously, it was reported that NMDAR stimulation in cortical neurons promotes, in a Ca2+-dependent manner, p35 cleavage to p25 by calpain leading to Cdk5 activation.20 Here, we show that, under our experimental culture conditions, rat primary cortical neurons efficiently responded to NMDAR stimulation, as judged by the MK801-inhibitable increased intracellular Ca2+ levels. Moreover, we show that glutamate induced Cdk5 activation in a process that was antagonized by MK801, indicating the involvement of NMDAR. Given that NMDAR-mediated activation of Cdk5 phosphorylates Cdh1 leading to APCCdh1 inhibition,17 we hypothesized that the stabilization of PFKFB3 could be consequence of NMDAR-mediated APCCdh1 inhibition. In good agreement with the presence of a nuclear-targeting motif in PFKFB3,21 we found that expressed PFKFB3 was localized in the nucleus, where neurons actively degraded it. In addition, we show that Cdh1 was phosphorylated by glutamate treatment, and that this was accompanied by cellular spread of PFKFB3 from the nucleus to cytosol in a Cdh1-inhibitable process; interestingly, the PFKFB3 mutant form lacking the Cdh1-recognizing KEN motif spontaneously accumulated. Together, these results indicate that PFKFB3 nuclear stabilization followed by cytosolic spread is the consequence of APCCdh1 inhibition. The mechanism whereby PFKFB3 is released from the nucleus remains unclear, although the physiological significance is likely in view of the cytoplasmic localization of the PFKFB3 target, 6-phosphofructo-1-kinase.
NMDAR-mediated PFKFB3 protein stabilization led to increased PFKFB3 activity and efficiently upregulated the rate of glycolysis in neurons. In previous studies, neurons failed to upregulate glycolysis immediately after the bioenergetic stress caused by mitochondrial inhibitors8, 22 or NMDAR activation.14 However, it should be noted that PFKFB3 stabilization takes place several hours after glutamate treatment, thus explaining the absence of measurable short-term glycolytic stimulation in cortical neurons in the previous studies.8, 14, 22 Accordingly, the delayed increase in glycolysis that we observe does not appear to be a neuronal attempt to rapidly compensate for the mitochondrial energy dysfunction, which occurs immediately after NMDAR stimulation.7 Instead, the delayed glycolysis activation reflects a long-term metabolic adaptation of neurons by excitotoxic insult; however, such an adaptation concurs with concomitant decrease in the rate of glucose oxidation through the PPP, hence, triggering oxidative stress and neurotoxicity. Intriguingly, although the stimulation of glycolysis is cytotoxic in neurons, it is cytoprotective in astrocytes.8 This different outcome shown by neurons and astrocytes is consistent with the expression of a robust antioxidant system in astrocytes that is not present in neurons.23 Upon inhibition of mitochondrial respiration, astrocytes switch on glycolysis, via the 5′-AMP-activated protein kinase-PFKFB3 pathway,14 to compensate for the ATP deficiency without affecting their antioxidant status.24 However, shifting glucose utilization from PPP to glycolysis in neurons compromises the efficacy of the critical antioxidant NADPH-glutathione regenerating system, hence causing delayed neurotoxicity. Importantly, both the increase in glycolysis and the decrease in PPP could be fully abolished by siPFKFB3, indicating that both metabolic pathways are wholly controlled by PFKFB3. In this context, it should be mentioned that Tp53-inducible glycolysis and apoptosis regulator (TIGAR), by catalyzing F2,6P2 degradation inhibits glycolysis and stimulates PPP.25 Thus, the control over F2,6P2 concentrations by either PFKFB3 – with a main fructose-6-phosphate-2-kinase activity12, 13 – or TIGAR – with fructose-2,6-bisphosphatase activity25 – appears to determine the fate of glucose metabolism. However, no evidence for NMDAR-mediated p53 upregulation is currently available, hence, remaining elusive whether TIGAR induction has a determinant role in neuronal metabolic change upon NMDAR stimulation.
The metabolic PPP to glycolysis shift triggered by NMDAR stimulation was accompanied by oxidative stress, as revealed both by an increase in the oxidized glutathione redox status and by the increased mitochondrial ROS, as well as apoptotic neuronal death. These data contrast with those reporting that NMDAR-mediated increase in neuronal ROS could be blocked with 6-aminonicotinamide, an inhibitor of PPP that produces NADPH required for NADPH oxidase activity.6 Whether the different neuronal settings (defined versus serum-based media) or the types of tools used (pharmacological versus genetic approaches to modulate glycolysis and PPP) are responsible for this apparent controversy remains elusive. However, in our hands, silencing PFKFB3 or PGI, which effectively inhibited glycolysis,15 or G6PD overexpression, a potent activator of the PPP,18 prevented such a metabolic switch and the concomitant ROS production by NMDAR stimulation. In fact, PPP activity produces reduced equivalents in the form of NADPH,26 which is also a necessary cofactor for antioxidant glutathione regeneration.18 Thus, PFKFB3 silencing significantly prevented the increase in oxidized glutathione status caused by NMDAR stimulation and this was critical at determining neuronal survival. Interestingly, it has been shown that, when oxidized, cytochrome c is released from mitochondria, hence promoting apoptotic neuronal death, and that PPP activity is essential at maintaining cytochrome c reduced.27 Our results, showing oxidative stress and neurodegeneration following PFKFB3 stabilization by NMDAR stimulation, confirm the critical role of PPP at regulating neuronal apoptosis. Furthermore, they show that the loss of PPP activity by APCCdh1 inhibition is a novel and important player in excitotoxicity. Together, these findings highlight the importance of metabolic modulation in excitotoxicity and neurodegeneration and emphasize that metabolic targets should be considered when designing therapeutic strategies.
Materials and Methods
Plasmid constructions and site-directed mutagenesis
The rat PFKFB3 full-length cDNA (splice variant K6; 1563 bp; accession number BAA21754) was obtained, by reverse-transcriptase polymerase chain reaction (RT-PCR), at our laboratory.15 PFKFB3 cDNA was fused, at its 5′-terminus, with the full-length cDNA encoding the GFP and subcloned in pCDNA3.0 vector. This GFP-PFKFB3 cDNA fusion construct was then subjected to site-directed mutagenesis of its KEN-box to AAA using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) using the following forward and reverse primers, respectively: 5′-ATCCTTCATTTTGCCGCAGCAGCTGACTTCAAGGC-3′ and 5′-ATGCCTTGAAGTCAGCTGCTGCGGCAAAATGAAGG-3′ (mutated nucleotides underlined). Human full-length Cdh1 cDNA (accession number NM_016263) was a generous gift of Dr. J Pines (Gurdon Institute, University of Cambridge, UK).
RNA interference
To knockdown PGI (accession number NM_207192), we used the following sequence for siRNA: 5′-CCTTACCAGACGTAGTGTT-3′ (nt 1248–1266). To knockdown PFKFB3 we used the sequence 5′-AAAGCCTCGCATCAACAGC-3′ (nt 1908–1926). Both siRNAs were previously validated at our laboratory for efficacy.14, 15 An siRNA against luciferase (5′-CTGACGCGGAATACTTCGATT-3′) was used as control.
RT-PCR analysis
Total RNA was purified from neurons using a commercially available kit (Sigma, Saint Louis, MO, USA). PFKFB3 mRNA expression was analyzed by 4.5% agarose electrophoresis after RT-PCR using the following forward and reverse oligonucleotides, respectively: 5′-CCAGCCTCTTGACCCTGATAAATG-3′ and 5′-TCCACACGCGGAGGTCCTTCAGAT-3′ for PFKFB3, and 5′-CTGGCGTCTTCACCACCAT-3′ and 5′-AGGGGCCATCCACAGTCTT-3′ for GAPDH. Reverse transcription was performed at 48 °C for 50 min, and PCR conditions were 10 min at 95 °C, 35 cycles of 1 min at 95 °C, 1 min at 58 °C and 30 s at 68 °C. Final extension was carried out for 10 min at 72 °C. In no case was a band detected by PCR without reverse transcription.
Antibodies
An anti-PFKFB3 (K3-K6 splice variants) antibody was generated, by rabbit immunization with the synthetic peptide 508MRSPRSGAESSQKH521-C, at our laboratory as previously described.15 A commercial anti-PFKFB3 antibody raised against a C-terminal region of the human PFKFB3 (protein accession Q16875; catalog number H00005209-M08, Novus Biologicals, Cambridge, UK) was also used; this antibody cross-reacts with human and rat PFKFB3, thus recognizing a region that is shared by all translational products of the rat K1 to K8 PFKFB3 mRNA splice variants. Anti-Cdh1 (AR38) was a generous gift from J Gannon (Clare Hall Laboratories, Cancer Research, UK). Anti-Cdk5 (C-8) and anti-PGI (K-16) were from Santa Cruz Biotechnology (Heidelberg, Germany). Anti-GFP was purchased from Abcam (Cambridge Science Park, Cambridge, UK). Anti-G6PD and anti-GAPDH were purchased from Sigma, and anti-phosphoserine from Zymed (Invitrogen, Groningen, The Netherlands).
Cell cultures
Cortical neurons in primary culture were prepared from fetal (E16) Wistar rats. Cells were seeded (2.5 × 105 cells/cm2) in DMEM (Sigma) supplemented with 10% (v/v) fetal calf serum (Roche Diagnostics, Heidelberg, Germany) and incubated at 37 °C in a humidified 5% CO2-containing atmosphere. After 48 h of plating, the medium was replaced with DMEM supplemented with 5% horse serum (Sigma) and with 20 mM D-glucose. On day 4, cytosine arabinoside (10 μM) was added in order to prevent non-neuronal proliferation. Cells were used by day 6, when enrichment was ∼99% (neurofilament; data not shown).
Cell treatments
Transfection of cells with plasmid vectors was carried out using 0.16–1.6 μg/ml of the plasmids, as indicated in the figure legends. All transfections were performed using lipofectamine 2000 (Invitrogen) following the manufacturer's instructions, at day 5 in vitro. After 6 h, the medium was removed and cells were further incubated overnight in the presence of culture medium. For RNA interference experiments, siRNAs (purchased from Thermo Fisher Scientific, Lafayette, CO, USA; sequences described above) were used. In dose-response preliminary settings, primary neurons were transfected, using Lipofectamine 2000 with 20–100 nM of the siRNAs, which showed a dose-dependent effect; only the results using 100 nM are shown. siRNA transfections were performed at day 3 in vitro and experiments were performed at day 6, when an efficient knockdown of the target proteins was obtained. For NMDAR activation, neurons at 6 days in vitro were incubated with 100 μM glutamate (plus 10 μM glycine) or 100 μM NMDA (plus 10 μM glycine) in buffered Hanks’ solution (pH 7.4) for 15 min.7 When indicated, incubations were performed in the presence of MK-801 (10 μM; Sigma). Neurons were then washed and further incubated in culture medium for the indicated time period.
Flow cytometric analysis of apoptotic cell death
APC/C-conjugated annexin-V and 7-aminoactinomycin D (7-AAD) (Becton Dickinson Biosciences, San Jose, CA, USA) were used to determine quantitatively the percentage of apoptotic neurons by flow cytometry. Cells were stained with annexin V-APC and 7-AAD, following the manufacturer's instructions, and were analyzed on a FACScalibur flow cytometer (15 mW argon ion laser tuned at 488 nm) using the CellQuest software software (BDB). Both GFP+ and GFP− cells were analyzed separately and the annexin V-APC-stained cells that were 7-AAD-negative were considered to be apoptotic.15
Detection of ROS
This was carried out using MitoSox-Red (Invitrogen). Neurons were incubated with 2 μM MitoSox-Red for 30 min, washed with PBS and the fluorescence assessed by flow cytometry.15
Measurement of the glycolytic and PPP fluxes
Suspensions of known amounts of cells (4–5 × 105 cells) obtained by smooth detaching from the cultures 6 h after glutamate treatments, were incubated in sealed vials containing a central well, which was used for 14CO2 or 3H2O trapping. Cells were incubated in the presence of 1 μCi of either D-[1-14C] glucose or D-[6-14C] glucose for PPP determinations, whereas 5 μCi of D-[3-3H] glucose were used for glycolytic flux determinations, both in a Krebs–Henseleit buffer (11 mM Na2HPO4, 122 mM NaCl, 3.1 mM KCl, 0.4 mM KH2PO4, 1.2 mM MgSO4, 1.3 mM CaCl2; pH 7.4) containing 5 mM D-glucose at 37 °C. In order to ensure an adequate O2 supply for oxidative metabolism by the cells throughout the 90 min incubation period, the gas phase in the vials containing the cells was supplied with extra O2 before the vials were sealed. The glycolytic flux was measured by assaying the rate of 3H2O production from [3-3H]glucose, as detailed previously.15 The PPP flux was measured by assessing the difference between 14CO2 production from [1-14C]glucose – which decarboxylates via the 6-phosphogluconate dehydrogenase-catalyzed reaction – and that of [6-14C]glucose – which decarboxylates via the tricarboxylic acid cycle.18, 28
Glutathione measurements
For glutathione determinations, neurons were treated with 1% (w/v) sulfosalicylic acid and centrifuged at 13 000 × g for 5 min at 4 °C. GSx (the amount of reduced glutathione (GSH), plus two times the amount of oxidized glutathione (GSSG)) and GSSG concentrations were measured in the supernatants using the enzymatic method of Tietze.29 GSSG was quantified after derivatization of GSH in the samples with 2-vinylpyridine. Data were extrapolated to those obtained with GSSG standards (0–5 μM for GSSG; 0–50 μM for GSx). The glutathione redox status was expressed as the GSSG/GSx ratio, as previously described.18, 30
Fura-2 fluorescence measurements
To estimate the intracellular Ca2+-dependent changes by NMDAR stimulation in cortical neurons we used the fluorescent probe Fura-2 (acetoxymethyl-derivative; Life Technologies, Eugene, OR, USA), as previously described.31 Essentially, neurons at 6 days in vitro, seeded in 96-well plates (Nunc), were incubated with Fura-2 (2 μM; dissolved in dimethyl sulphoxide (DMSO)) for 40 min in DMEM at 37 °C. Then, cells were washed and further incubated with standard buffer (140 mM NaCl, 2.5 mM KCl, 15 mM Tris-HCl, 5 mM D-glucose, 1.2 mM Na2HPO4, 1 mM MgSO4 and 1 mM CaCl2, pH 7.4) for 30 min and 37 °C. Finally, the standard buffer was removed and experimental buffer (140 mM NaCl, 2.5 mM KCl, 15 mM Tris-HCl, D-glucose, 1.2 mM Na2HPO4, and 2 mM CaCl2, pH 7.4), either in the absence or in the presence of MK801 (10 μM), was added. Emissions at 510 nm, after excitations at 335 and 363 nm, respectively, were recorded at 1 s intervals in a Varioskan Flash (Thermo Fischer, Vantaa, Finland) spectrofluorometer at 32 °C. After ∼10 s, glutamate (100 μM) or NMDA (100 μM) (plus 10 μM glycine) was injected and emissions were further recorded for 50 s. Ca2+-dependent fluorescence changes were estimated by representing the ratio of fluorescence emitted at 510 nm obtained after excitation at 335 nm divided by that at 363 nm (F335/F363). Background subtraction was accomplished from emission values obtained in Fura-2-lacking (DMSO-containing) neurons. In preliminary experiments, the Ca2+ specificity of the measurements was tested in Ca2+-free experimental buffer containing 1 mM ethylene glycol tetraacetic acid (EGTA), which fully prevented the changes in 510 nm emissions (data not shown). At least, six wells were recorded per condition in each experiment (n=4 experiments) and the averaged values are shown.
Western blot analysis
After transfections and treatments, neurons were lysed in RIPA buffer (2% sodium dodecylsulphate, 2 mM ethylene diamine tetraacetic acid (EDTA), 2 mM EGTA, 50 mM Tris; pH 7.5), supplemented with phosphatase inhibitors (1 mM Na3VO4, 50 mM NaF) and protease inhibitors (100 μM phenylmethylsulfonyl fluoride, 50 μg/ml anti-papain, 50 μg/ml pepstatin, 50 μg/ml amastatin, 50 μg/ml leupeptin, 50 μg/ml bestatin and 50 μg/ml soybean trypsin inhibitor) and boiled for 5 min. Aliquots of cell extracts were subjected to sodium dodecyl sulfate (SDS) polyacrylamide gel (MiniProtean, Bio-Rad, Hercules, CA, USA) and blotted with antibodies overnight at 4 °C. Signal detection was performed with an enhanced chemiluminescence kit (Pierce, Thermo Scientific, Waltham, MA, USA).
Immunoprecipitation and Cdk5 kinase activity
Neurons were lysed in ice-cold buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10 mM EDTA, 2 mM EGTA, 1% NP-40, supplemented with the phosphatase and protease inhibitors cited above. Cell extracts were clarified by centrifugation and supernatants (50 μg of protein for immunoprecipitation experiments, 500 μg for Cdk5 kinase assays) were incubated with anti-Cdh1 or anti-Cdk5, overnight at 4 °C, followed by the addition of 15–30 μl of protein A-sepharose (GE Healthcare Life Sciences, Uppsala, Sweden) for 1–2 h at 4 °C. Immunoprecipitates were extensively washed with lysis buffer and either detected by western blot analysis against anti-phosphoserine or resuspended in kinase buffer (20 mM Tris-HCl pH 7.6, 20 mM MgCl2, 2 mM MnCl2, 1 mM EDTA, 1 mM EGTA, 0.1 mM dithiothreitol) containing 20 μM ATP, 10 μCi of [γ-32P]ATP and histone-H1 (50 μg/ml; Sigma) for SDS-polyacrylamide gel (12%) electrophoresis; transferred proteins were visualized by autoradiography and anti-Cdk5 blotting.32
Protein determinations
Protein concentrations were determined in the cell suspensions, lysates or in parallel cell culture incubations after solubilization with 0.1 M NaOH. Protein concentrations were determined as described33 using bovine serum albumin as a standard.
Confocal microscopy
Neurons were grown on glass coverslips. After transfections and treatments they were fixed with 4% (v/v in PBS) paraformaldehyde for 20 min and incubated with DAPI (30 μM; Sigma). Confocal microscopy images were obtained with a Leica SP5 microscope (DMI-6000B model; Leica Microsystems GmbH, Wetzlar, Germany).
Statistical analysis
Measurements from individual cultures were always carried out in triplicate. The results are expressed as mean±S.E.M. values for three different culture preparations. Statistical analysis of the results was performed by one-way analysis of variance (ANOVA), followed by the least significant difference multiple range test. In all cases, P<0.05 was considered significant.
Abbreviations
- ANOVA:
-
analysis of variance
- APC:
-
anaphase-promoting complex
- Cdk5:
-
cyclin-dependent kinase-5
- cDNA:
-
complementary DNA
- DMSO:
-
dimethyl sulphoxide
- EDTA:
-
ethylene diamine tetraacetic acid
- EGTA:
-
ethylene glycol tetraacetic acid
- G6PD:
-
glucose-6-phosphate dehydrogenase
- GFP:
-
green fluorescent protein
- GSH:
-
reduced glutathione
- GSSG:
-
oxidized glutathione
- GSx:
-
total glutathione
- NMDA:
-
N-methyl-D-aspartic acid
- NMDAR:
-
N-methyl-D-aspartic acid receptor
- PFKFB3:
-
6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3
- PGI:
-
phosphoglucose isomerase
- PPP:
-
pentose-phosphate pathway
- ROS:
-
reactive oxygen species
- SDS:
-
sodium dodecyl sulfate
- siRNA:
-
small interfering RNA
- TIGAR:
-
Tp53-inducible glycolysis and apoptosis factor
References
Papadia S, Soriano FX, Leveille F, Martel MA, Dakin KA, Hansen HH et al. Synaptic NMDA receptor activity boosts intrinsic antioxidant defenses. Nat Neurosci 2008; 11: 476–487.
Hardingham GE, Bading H . Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci 2010; 11: 682–696.
Bossy-Wetzel E, Schwarzenbacher R, Lipton SA . Molecular pathways to neurodegeneration. Nat Med 2004; 10: S2–S9.
Bano D, Young KW, Guerin CJ, Lefeuvre R, Rothwell NJ, Naldini L et al. Cleavage of the plasma membrane Na+/Ca2+ exchanger in excitotoxicity. Cell 2005; 120: 275–285.
Celsi F, Pizzo P, Brini M, Leo S, Fotino C, Pinton P et al. Mitochondria, calcium and cell death: a deadly triad in neurodegeneration. Biochim Biophys Acta 2009; 1787: 335–344.
Brennan AM, Suh SW, Won SJ, Narasimhan P, Kauppinen TM, Lee H et al. NADPH oxidase is the primary source of superoxide induced by NMDA receptor activation. Nat Neurosci 2009; 12: 857–863.
Almeida A, Bolaños JP . A transient inhibition of mitochondrial ATP synthesis by nitric oxide synthase activation triggered apoptosis in primary cortical neurons. J Neurochem 2001; 77: 676–690.
Almeida A, Almeida J, Bolaños JP, Moncada S . Different responses of astrocytes and neurons to nitric oxide: the role of glycolytically-generated ATP in astrocyte protection. Proc Natl Acad Sci USA 2001; 98: 15294–15299.
Bolaños JP, Almeida A, Moncada S . Glycolysis: a bioenergetic or a survival pathway? Trends Biochem Sci 2010; 35: 145–149.
Allaman I, Belanger M, Magistretti PJ . Astrocyte-neuron metabolic relationships: for better and for worse. Trends Neurosci 2011; 34: 76–87.
Pellerin L . Food for thought: the importance of glucose and other energy substrates for sustaining brain function under varying levels of activity. Diabetes Metab 2010; 36 (Suppl 3): S59–S63.
Manzano A, Rosa JL, Ventura F, Perez JX, Nadal M, Estivill X et al. Molecular cloning, expression, and chromosomal localization of a ubiquitously expressed human 6-phosphofructo-2-kinase/ fructose-2, 6-bisphosphatase gene (PFKFB3). Cytogenet Cell Genet 1998; 83: 214–217.
Hirata T, Kato M, Okamura N, Fukasawa M, Sakakibara R . Expression of human placental-type 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatase in various cells and cell lines. Biochem Biophys Res Commun 1998; 242: 680–684.
Almeida A, Moncada S, Bolaños JP . Nitric oxide switches on glycolysis through the AMP protein kinase and 6-phosphofructo-2-kinase pathway. Nat Cell Biol 2004; 6: 45–51.
Herrero-Mendez A, Almeida A, Fernandez E, Maestre C, Moncada S, Bolaños JP . The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat Cell Biol 2009; 11: 747–752.
Almeida A, Bolaños JP, Moreno S . Cdh1/Hct1-APC is essential for the survival of postmitotic neurons. J Neurosci 2005; 25: 8115–8121.
Maestre C, Delgado-Esteban M, Gomez-Sanchez JC, Bolaños JP, Almeida A . Cdk5 phosphorylates Cdh1 and modulates cyclin B1 stability in excitotoxicity. Embo J 2008; 27: 2736–2745.
Garcia-Nogales P, Almeida A, Bolaños JP . Peroxynitrite protects neurons against nitric oxide-mediated apoptosis. A key role for glucose-6-phosphate dehydrogenase activity in neuroprotection. J Biol Chem 2003; 278: 864–874.
Delgado-Esteban M, Almeida A, Bolaños JP . D-Glucose prevents glutathione oxidation and mitochondrial damage after glutamate receptor stimulation in rat cortical primary neurons. J Neurochem 2000; 75: 1618–1624.
Lee MS, Kwon YT, Li M, Peng J, Friedlander RM, Tsai LH . Neurotoxicity induces cleavage of p35 to p25 by calpain. Nature 2000; 405: 360–364.
Yalcin A, Clem BF, Simmons A, Lane A, Nelson K, Clem AL et al. Nuclear targeting of 6-phosphofructo-2-kinase (PFKFB3) increases proliferation via cyclin-dependent kinases. J Biol Chem 2009; 284: 24223–24232.
Bolaños JP, Heales SJR, Land JM, Clark JB . Effect of peroxynitrite on the mitochondrial respiratory chain: differential susceptibility of neurones and astrocytes in primary cultures. J Neurochem 1995; 64: 1965–1972.
Makar TK, Nedergaard M, Preuss A, Gelbard AS, Perumal AS, Cooper AJL . Vitamin E, ascorbate, glutathione, glutathione disulfide, and enzymes of glutathione metabolism in cultures of chick astrocytes and neurones: evidence that astrocytes play an important role in antioxidative processes in the brain. J Neurochem 1994; 62: 45–53.
Bolaños JP, Peuchen S, Heales SJR, Land JM, Clark JB . Nitric oxide-mediated inhibition of the mitochondrial respiratory chain in cultured astrocytes. J Neurochem 1994; 63: 910–916.
Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 2006; 126: 107–120.
Hothersall JS, Baquer NZ, Greenbaum AL, McLean P . Alternative pathways of glucose utilization in brain. Changes in the pattern of glucose utilization in brain during development and the effect of phenazine methosulphate on the integration of metabolic routes. Arch Biochem Biophys 1979; 198: 478–492.
Vaughn AE, Deshmukh M . Glucose metabolism inhibits apoptosis in neurons and cancer cells by redox inactivation of cytochrome c. Nat Cell Biol 2008; 10: 1477–1483.
Larrabee MG . Evaluation of the pentose phosphate pathway from 14CO2 data. Fallibility of a classic equation when applied to non-homogeneous tissues. Biochem J 1990; 272: 127–132.
Tietze F . Enzyme method for quantitative determination of nanogram amounts of total and oxidized glutathione: application to mammalian blood and other tissues. Anal Biochem 1969; 27: 502–522.
Dringen R, Pfeiffer B, Hamprecht B . Synthesis of the antioxidant glutathione in neurons: supply by astrocytes of CysGly as precursor for neuronal glutathione. J Neurosci 1999; 19: 562–569.
Ring A, Tanso R . Measurements with fluorescent probes in primary neural cultures; improved multiwell techniques. J Pharmacol Toxicol Methods 2007; 56: 300–307.
Wang J, Liu S, Fu Y, Wang JH, Lu Y . Cdk5 activation induces hippocampal CA1 cell death by directly phosphorylating NMDA receptors. Nat Neurosci 2003; 6: 1039–1047.
Lowry OH, Rosebrough NJ, Lewis-Farr A, Randall RJ . Protein measurement with the Folin phenol reagent. J Biol Chem 1951; 193: 265–275.
Acknowledgements
The technical assistance of Monica Resch is gratefully acknowledged. This work was funded by the Spanish Ministerio de Ciencia e Innovacion (Consolider-Ingenio CSD2007-00020; SAF2010-20008), Instituto de Salud Carlos III (PS09/0366), FEDER (European regional development fund) and the Junta de Castilla y Leon (GREX206). PR-R is a recipient of a predoctoral FPU fellowship from the Ministerio de Ciencia e Innovación.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Deshmukh
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Rodriguez-Rodriguez, P., Fernandez, E., Almeida, A. et al. Excitotoxic stimulus stabilizes PFKFB3 causing pentose-phosphate pathway to glycolysis switch and neurodegeneration. Cell Death Differ 19, 1582–1589 (2012). https://doi.org/10.1038/cdd.2012.33
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2012.33
Keywords
This article is cited by
-
Deficits in mitochondrial TCA cycle and OXPHOS precede rod photoreceptor degeneration during chronic HIF activation
Molecular Neurodegeneration (2023)
-
Axonal energy metabolism, and the effects in aging and neurodegenerative diseases
Molecular Neurodegeneration (2023)
-
Klotho increases antioxidant defenses in astrocytes and ubiquitin–proteasome activity in neurons
Scientific Reports (2023)
-
Exploring the Role of Glycolytic Enzymes PFKFB3 and GAPDH in the Modulation of Aβ and Neurodegeneration and Their Potential of Therapeutic Targets in Alzheimer’s Disease
Applied Biochemistry and Biotechnology (2023)
-
Treatment against glucose-dependent cancers through metabolic PFKFB3 targeting of glycolytic flux
Cancer and Metastasis Reviews (2022)