Abstract
Autophagy is an evolutionarily conserved catabolic process that involves the invagination and degradation of cytoplasmic components through an autophagosomelysosome track. Autophagy functions as a quality control of cellular milieu and is implicated in a wide variety of pathological conditions. However, excessive or imbalanced autophagic flux may also be associated with cellular toxicity and may potentially contribute to the development of pathological conditions. Just as all membrane trafficking systems need to constantly strike a balance in their level of activation and inhibition to ensure proper spatial and temporal delivery of their cargo, autophagy must also be tightly regulated. Here, we provide an overview of the current knowledge regarding the negative regulation of mammalian autophagy in an effort to understand its physiological relevance and potential clinical importance.
Similar content being viewed by others
Main
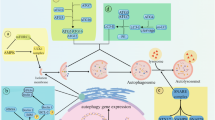
Autophagy, originally described as a lysosome-dependent bulk degradation of cytoplasmic components on starvation, has since been shown to influence diverse aspects of homeostasis and is implicated in many diseases.1 The autophagic cascade is initiated by the engulfment of cytoplasmic cargoes, including long-lived or aggregated proteins, defective organelles, and various soluble molecules, by a double-membraned autophagosome. The autophagosome is then progressively acidified by fusion with the late endosome and the lysosomal compartment to form the autolysosome, exposing the inner compartment to lysosomal hydrolases. Eventually, the inner membrane of the autophagosome and its sequestered contents are degraded, with the resulting macromolecules recycled (Figure 1).1, 2 Over 30 distinct autophagy-related (Atg) genes have been identified so far in yeast.3 Many of the genes have their orthologs in mammals.1, 3Classical autophagy can be largely divided into several major steps that require specific Atg genes.1 These include (a) target of rapamycin (TOR)-mediated induction of autophagy through an Atg1-kinase complex, (b) initiation of autophagosome formation through a Beclin1-associated class III phosphoinositide (PI) 3-kinase (PI3KC3) complex, (c) autophagosome membrane elongation and completion, involving two evolutionarily conserved ubiquitin-like conjugation systems–the Atg12–Atg5 and the LC3 (the mammalian ortholog of yeast Atg8)-PE (phosphatidylethanolamine) systems, (d) autophagosome maturation through fusion with lysosomes, and (e) cargo degradation through lysosomal enzymes and recycling (Figure 1).1 It is important to note that it is the combined signaling cascades that regulate autophagy, coupled with the concerted action of these distinct steps, that lead to the overall cellular autophagic response.
Negative regulation of the autophagy pathway. Schematic representation of the autophagy pathway and its negative regulation in mammalian cells. Autophagy proceeds through a series of steps including signal transduction, membrane nucleation, elongation, and completion of the autophagosome, autophagosome maturation by fusion with a lysosome, followed by degradation of autophagic cargoes, and recycling of the resulting molecules. Negative regulation occurs at each dynamic step. Autophagy induction is directly and tightly controlled by the ‘nutrient sensor’ mTOR, which senses and integrates signals from numerous sources including growth factors, amino acids, hypoxia, and energy levels. Under nutrient-rich conditions, mTOR associates with the ULK1 complex comprised of ULK1 (mammalian Atg1), mammalian Atg13 (mAtg13), and FIP200 (mammalian Atg17), and phophorylates ULK1 and mAtg13 to repress autophagy. On inactivation of mTOR by upstream signaling, it dissociates from the ULK1 complex, triggering the autophagy cascade. Autophagosome nucleation is confined to phosphatidylinositol-3phosphate (PtdIns(3)P)-containing vesicles and is driven by the Beclin1-associated PI3KC3 complex. The antiapoptotic Bcl-2 and its interactor, NAF-1, negatively regulate autophagy by sequestering Beclin1 from the complex, thereby restricting the production of PtdIns(3)-P. Similarly, the PtdIns(3)-P phosphatase, Jumpy, catalyzes the reverse reaction of PtdIns(3)-P hydrolysis to inhibit autophagy. Beclin1 can also be cleaved by caspases, which destroys its proautophagic activity. Two ubiquitin-like conjugation systems are involved in autophagosomal membrane expansion and completion: one is LC3–phosphatidylethanolamine (PE) conjugation and the other is Atg12–Atg5 conjugation. The interaction of LC3 with the E2-like enzyme, Atg3, is inhibited by FLIP, whereas Atg5 cleavage by calpain attenuates Atg5–Atg12 conjugation, resulting in apoptosis through the formation of a complex with Bcl-xL in the mitochondria. Autophagosome fusion with the lysosome signifies the maturation stage of autophagy, a step that is least understood but found to be suppressed by the Rubicon complex (Rubicon–UVRAG–Beclin1–PI3KC3–p150). Finally, the end products of proteolysis suppress autophagy by upregulating mTOR signaling, serving as a negative feedback loop
Recently, an alternative autophagy pathway inducible by cellular stress was discovered in mouse embryonic fibroblasts (MEFs) depleted of the key autophagosomal proteins, Atg5 and Atg7.4 Interestingly, this Atg5- and Atg7-independent autophagy pathway is not associated with LC3 lipidation but seems to specifically involve membranes derived from trans-Golgi and late endosomes.4 Thus, autophagy seems to be far more complex than was previously realized, demanding sophisticated autophagosomal sorting, conceivably encompassing multiple routes of trafficking while orchestrating regulation of membrane dynamics.1, 5
The activation of autophagy is a double-edged sword. On the one hand, autophagy is essential for provoking a protective response and enhancing the cells’ adaptation to metabolic stresses and immunological challenges.6, 7 In particular, infection, inflammation, neurodegeneration, and cancer can result from pathogen-induced or inherited disruption of the autophagy pathway.6, 8 On the other hand, excessive autophagic activation, derailed autophagic trafficking, or imbalanced degradation and recycling can give rise to pathogenic conditions by straining critical cellular constituents, leading to cell degeneration and toxicity.9 An example is cardiac myocyte death during ischemia/reperfusion, which involves excessively stimulated autophagy and can be alleviated with treatment of 3-methyladenine, an autophagy inhibitor, or knockdown of beclin1, an essential autophagy effector.10, 11 Moreover, although autophagy has been found to retard cardiac hypertrophy by increasing protein degradation, prolonged autophagy seems to have a crucial part in the transition from cardiac hypertrophy to decompensated heart failure, likely because of loss of cardiac mass.11 Indeed, overwhelming autophagic turnover or the build-up of a ‘traffic jam’ in autophagic vacuoles predominates the pathology of many types of myopathy.12 In such conditions, autophagy is not simply a by-product of the cells’ recovery program, but rather a phenomenon that can directly contribute to cell pathologies. Similarly, neuronal atrophy, neurite degeneration, and cell death can be exacerbated by the upregulation of autophagy, but are partially reversed by the inactivation of some essential autophagy genes, at least in certain contexts.13 Autophagy may also facilitate pathogenesis of infections.14 Prominent examples include poliovirus, which intercepts autophagy to provide a membrane scaffold for viral replication,15 and HIV, which hijacks autophagy as a bystander death program for CD4+ T-cell depletion and immunodeficiency.16 Although it is still a matter of debate whether autophagy is ultimately a protective response or a detrimental process, it seems clear that, if left unchecked, autophagy can lead to adverse effects in cells under certain pathological conditions. Therefore, autophagy signaling and functions are necessities, but ones that must be under tight negative regulation.
A series of negative regulators of autophagy have been identified in the past 10 years (Table 1), and a picture of how the balance of autophagy in mammals is achieved is beginning to emerge. It should be noted that these negative regulators do not work as a single entity, but are orchestrated to function at multiple levels within the autophagy cascade. The timing of autophagy induction is stringently controlled by the nutrient sensor, the mammalian TOR (mTOR) kinase, which ensures the faithful application of autophagy in the appropriate physiological setting by preventing its direct activation.17 Once autophagy is provoked, the autophagy flux can be further restrained at each stage of membrane dynamics: nucleation, elongation, maturation, and degradation.2
The overall magnitude of the entire autophagic ‘wave’ is determined by the activities associated with each of the steps illustrated in Figure 1. The end products of autophagy can serve as a negative-feedback regulatory mechanism to preclude prolonged activity and to synchronize the positive and negative regulation of signal transduction.18 Moreover, the post-translational modification of key autophagosomal proteins provides additional forms of regulation.19, 20, 21 Understanding how autophagy is regulated is important, as it will facilitate the development of new strategies for the control of autophagy-associated pathologies. As many reviews have discussed the positive regulation of autophagy,22 and as recently negative regulation of autophagy has been intensively studied, we herein summarize our current knowledge of the negative regulation of mammalian autophagy and discuss its physiological relevance and potential clinical importance (also shown in Figure 1).
mTOR: the gatekeeper of autophagy signaling
The kinase mTOR of the phosphoinositide 3-kinase-related kinase (PIKK) family serves as a key signaling ‘station’ in the regulation of cellular metabolism, which promotes protein synthesis and suppresses the induction of autophagy.23 mTOR is found to be part of at least two functionally distinct complexes: the rapamycin-sensitive mTOR complex 1 (mTORC1) and the rapamycin-insensitive mTOR complex 2 (mTORC2). mTORC1 contains mTOR, along with the subunits regulatory-associated protein of mTOR (Raptor), lethal with SEC13 protein 8 (LST8), DEPTOR, and proline-rich AKT-substrate (PRAS40). This complex integrates a plethora of signaling pathways that respond to nutrition status and growth factors.24 The second complex, mTORC2, contains mTOR along with the subunits rapamycin-insensitive companion of mTOR (Rictor), stress-activated-protein-kinase-interacting protein 1 (SIN1), LST8, and DEPTOR. This complex is less understood, but has been shown to be involved in the regulation of actin organization and activation of AKT.25
When nutrients are plentiful, the PI3K-Akt signaling pathway, stimulated by growth factors, affects mTORC1 (Figure 1).24 This triggers a cascade of anabolic processes for cell growth and proliferation, mainly mediated by the ribosomal S6 kinase (p70S6 K) and eukaryotic translation initiation factor 4E (eIF4E)-binding protein (4E-BP), two direct downstream substrates of mTORC1.24 Concomitantly, activation of mTORC1 leads to autophagy suppression.23 This is achieved by direct inhibition of the Atg1Atg13–Atg17 complex, which is required for the initiation of autophagosome formation in both yeast and mammals.23, 26 In yeast, TOR activation leads directly to phosphorylation of Atg13, precluding its interaction with Atg1 to thereby inhibit the biogenesis of autophagosomes.27 In support of this view, expression of an unphosphorylatable form of Atg13 induces autophagy regardless of TOR activation.27 In mammals, the analogous mTORC1–Atg1 complex axis shows slightly different properties: mTOR interacts with the mammalian Atg1 complex, the ULK1 complex, and phosphorylates ULK1 and mammalian Atg13 to inhibit autophagy, whereas dissociation of mTORC1 leads to activation of ULK1 (Figure 1).28, 29 As activation of the Atg1/ULK1 complex is essential for most, if not all, cases of autophagy initiation, induction of autophagy can occur autonomously in the absence of any exogenous or endogenous stimuli when mTORC1 is inactivated or inhibited, as occurs with rapamycin treatment.30 Therefore, mTOR may constitute an early means of limiting autophagy signaling in favor of maximizing the synthesis of proteins, lipids, and nucleic acids for cell growth, most notably in conditions of nutrient availability.24 Under conditions of low energy or hypoxia, mTORC1 is repressed by AMP-activated protein kinase (AMPK), which responds to an increased AMP/ATP ratio under low-energy conditions, and REDD1, a transcriptional target of HIFα that is stimulated by hypoxia.23, 31 Thus, mTORC1 inactivation releases the Atg1/ULK1 complex from a repressed state, leading to the stimulation of autophagy.32 In this context, autophagy-mediated breakdown and recycling of cellular components becomes the preferred route of energy production.
On the basis of the central role of mTORC1 in regulating cell growth and proliferation, it is perhaps unsurprising that metabolic signaling upstream of mTORC1 seems profoundly altered in many tumors.33 The oncogenic components in the PI3K–AKTmTOR-mediated anabolic pathway are constitutively active in most cancers, whereas molecules known to suppress mTORC1, including PTEN, AMPK, TSC1/TSC2, and p53, are often mutated.34 Accordingly, the mTORC1 inhibitor, rapamycin, and its analogs are actively pursued as a treatment for human malignancies. Despite gaps in our understanding, it is already clear that mTORC1 signaling and its manipulation of the autophagic process represent plausible targets for cancer therapy.
Phosphatidylinositol-3-phosphate (PtdIns(3)P): a spatial control of autophagy nucleation
Following autophagy induction, signals originating from mTORC1 are resolved in space, to a membrane nucleation site rich in PtdIns(3)P. Recent studies by Axe et al.35 have shown that these PtdIns(3)P-enriched structures (denoted omegasomes because of their Ωlike shape) seem generated at the ER membrane, which provides a docking site for assembly of the autophagy machinery. Subsequent study by Halley et al.36 suggested that the outer membranes of mitochondria in mammalian cells can serve as an alternative origin of the autophagosomal membranes, in particular, during starvation-induced autophagy, by providing PE, the lipid target of the autophagosome marker LC3, highlighting multiple sites of autophagosome nucleation and an intimate connection between ER and mitochondria in autophagy. Notably, production of PtdIns(3)P is mainly governed by the Beclin1-associated PI3KC3 complex, whereas PtdIns(3)P phosphatases mediate PtdIns(3)P hydrolysis to maintain a balanced reservoir of PtdIns(3)P in the cell.37 In addition, there is a level of inhibitory control over the function of the Beclin1–PI3KC3 complex that links the production of PtdIns(3)P with cell survival and apoptosis regulation, exemplified by the roles of Bcl-2 and the recently identified nutrient-deprivation autophagy factor-1 (NAF-1) in autophagy regulation.38, 39 The level of PtdIns(3)P in a cell must be carefully maintained, as the subversion of PtdIns(3)P homeostasis may be a fundamental mechanism in the development of congenital myopathies.37
Jumpy
Jumpy, also known as MTMR14, encodes a PtdIns(3)P phosphatase that belongs to the myotubularin (MTM) family. This family contains 15 members of lipid phosphatases characterized by a conserved protein tyrosine phosphatase (PTP) domain, with specificity for PtdIns(3)P and PtdIns(3,5)P2 in mammalian cells.40 In a siRNA-based screen of MTM family members for autophagy regulation, Vergne et al.41 found that depletion of jumpy, but not of other MTM members, significantly increases both basal and starvation-induced autophagy. Furthermore, Jumpy associates with isolation membranes and promotes the PtdIns(3)P-dependent recruitment of the early autophagic protein, WIP-1(mammalian Atg18). In contrast, the overexpression of Jumpy delays traffic out of the autophagosomal compartment.41 Although the details remain hazy, these results show that Jumpy is an effective negative regulator of autophagy that drives the reverse reaction of PtdIns(3)P hydrolysis to control excessive PtdIns(3)P-mediated signaling. In vivo, jumpy-deficient mice display muscle weakness and fatigue, a phenotype similar to that seen in patients with centronuclear myopathy (CNM, a severe congenital muscular disorder characterized by prominent myonuclei internalization and centralization).42 Consistent with this, Jumpy and another myotubularin, MTM1, have been genetically linked to CNM.43, 44 Notably, siRNA knockdown of MTM1 in myocytes fails to give rise to enhanced autophagy, as seen with jumpy,41 suggesting that the subversion of the autophagosomal PtdIns(3)P balance does not explain all cases of CNM, and disparate pathomechanisms might be responsible for similar clinical features of CNM. As PtdIns(3)P homeostasis is also critical for endosome trafficking and Ca2+ metabolism, whether Jumpy-associated myopathies are solely due to an overwhelmed autophagic self-consumption and/or aberrant endosome function remains to be tested. Nevertheless, expression of the R336Q missense mutant of Jumpy found in CNM patients completely abrogates Jumpy phosphatase activity and results in an increased level of autophagy in vitro, indicating that aberrant recycling of autophagosomal PtdIns(3)P may contribute, at least in part, to disease etiologies.44 Recently, MTMR3 has been found to regulate constitutive autophagy initiation and the size of autophagic vacuoles in epithelial cells, favoring the idea that autophagosomal PtdIns(3)P regulation might be a shared property of the MTM family.37
Bcl-2 and its assistant, NAF-1
Unlike Jumpy, which promotes the PtdIns(3)P hydrolysis to negatively regulate autophagy, antiapoptotic Bcl-2 proteins such as Bcl-2, Bcl-xL, and Bcl-w inhibit autophagy by targeting the ER-associated PI3KC3 complex to restrict PtdIns(3)P production.38, 45 The core PI3KC3 complex, including Beclin1, PI3KC3, and p150, exists in at least three configurations: the Atg14 complex, which contains an additional subunit of Atg14, functions at the early stage of autophagosome formation;26, 46 the UVRAG-Bif-1 complex, which has UVRAG and Bif-1 (also called endophilin B1) instead, facilitates membrane curvature of autophagosomes;26, 47, 48 and the Rubicon complex, which harbors Rubicon and UVRAG, functions in autophagosome maturation as discussed below.26, 46, 49 Although the specific function of each individual subunit remains to be established, Beclin1 expression and its interaction with PI3KC3 have been shown to be indispensable for autophagosome formation.50, 51 Antiapoptotic Bcl-2, Bcl-xL, and Bcl-w bind and hold Beclin1 at bay, thereby preventing Beclin1 from assembling the autophagy-inducing PI3KC3 complex.38, 50, 51 Consistent with this view, ER-localized Bcl-2, but not mitochondrial-localized Bcl-2, confers autophagy inhibition, most notably when nutrients are readily available.38 After stimulation with stress, the negative regulation of Bcl-2 can be overcome by contextually different mechanisms, including the phosphorylation of Bcl-2 or Beclin1 by JNK or DAP-kinase (DAPk), respectively. In addition, the competitive binding of BH3-only proteins (e.g., Bad) or pharmacological BH3 mimetic agents (e.g., ABT737) to Bcl-2 releases Beclin1 from Bcl2 inhibition.51, 52 Indeed, a Beclin1 mutant that no longer binds to Bcl-2 induces a detrimental autophagy response.38 Thus, the status of the Bcl-2–Beclin1 interaction might be a switch for the Beclin1–PI3KC3-mediated autophagic response in the ER.
Shore et al. have recently added another twist to the Bcl-2–Beclin1 crosstalk by showing that ER-associated NAF-1 (CISD2: CDGSH iron sulfur domain 2; synonyms: ZCD2, Noxp70, and Miner1) is required for Bcl-2–Beclin1 interaction and Bcl-2 inhibition of autophagy at the ER.53 Knockdown of NAF-1 diminishes the Bcl-2 binding to Beclin1 and triggers autophagy.50, 53 Unlike Beclin1, NAF-1 does not engage the conventional hydrophobic BH3-binding pocket on the Bcl-2 surface for binding. Moreover, the ER-restricted proapoptotic BH3-only protein, Bik, can displace NAF-1 from Bcl-2 and induce autophagy.50, 53 A plausible explanation of this finding is that NAF1 might bind to a hitherto unknown site on Bcl-2 to facilitate a conformational change needed for an efficient Beclin1 interaction. This may be reversed by Bik because of its competitive affinity for Bcl-2. Of special note, depletion of NAF-1 seems to have a minimal effect on the steady-state level of autophagy, but autophagy is strongly evoked in starved cells with an unusual distribution of LC3 at membrane blebs and filopodia.53 This indicates that the loss of NAF-1 might influence both the magnitude and spatial control of Beclin1-mediated autophagy flux. Consistently, Cisd2 (naf-1) deficiency in mice causes autophagic cell death in multiple tissues accompanied by a panel of premature aging phenotypes.54
Similar to Bcl-2,55 NAF-1 is found in physical association with the inositol 1,4,5trisphosphate (IP3) receptor (IP3R), an IP3 sensor and an ER-Ca2+ efflux channel, and is required for the Bcl-2-mediated depression of ER Ca2+ store.53 It should be noted that IP3R also regulates autophagy and that the antagonists (e.g., xestospongins) of IP3R strongly induce autophagy.56 Intriguingly, such an effect of IP3R on autophagy seems to be irrelevant to its regulation of intracellular Ca2+ levels but is dependent on Beclin1 interaction.57 Whether the dual roles of NAF-1 in Bcl-2-mediated Beclin1 inhibition and IP3 receptor activity represent two sides of the same coin or are functionally independent regulatory mechanisms remains to be further tested. Nevertheless, the coordinated regulation of autophagic flux with Ca2+ metabolism by the NAF-1/Bcl-2 complex and IP3 receptor integrates autophagy into the broader cellular signaling network. Not surprisingly, tumors exploit these mechanisms to gain a selective advantage. For example, cancer cells often express more Bcl-2 to prolong the inhibition of apoptosis and autophagy to ensure cancer cell proliferation, in much the same way as herpesviruses, such as γ-Herpesvirus 68, maintain their latent infections in host cells.52, 58
Recent study by Furuya et al.59 adds another piece to the regulation of the PtdIns(3)P production, showing that cycline-dependent kinases (Cdks) including Cdk1 and Cdk5 can phosphorylate PI3KC3 on Thr159, which inhibits PI3KC3 interaction with Beclin1 and negatively regulates autophagy. This study thus provides a mechanism by which autophagy is controlled in cell cycle progression.
FLIP (FLICE-like inhibitory protein) and cleaved Atg5: brakes for membrane elongation
Autophagosome membrane elongation involves two ubiquitin-like conjugation systems: LC3 (mammalian Atg8)-PE (phophatidylethanolamine) conjugation and Atg12–Atg5 conjugation.60 After proteolytic cleavage of its carboxy terminus by the Atg4 cysteine protease, LC3 is conjugated to the membrane lipid PE after sequential processing by Atg7 (E1-like enzyme) and Atg3 (E2-like enzyme).60 In this conjugation process, FLIP tunes the interaction between ubiquitin-like LC3 and Atg3.61 Meanwhile, Atg12 is conjugated to Atg5 in a similar manner, except that Atg10 instead of Atg3 is used as the E2 enzyme. Furthermore, under the control of calpain-mediated cleavage, the membrane elongation factor, Atg5, can switch from an autophagy mode to an apoptosis mode on apoptotic stimulation.19
FLIP's flipping autophagy
Cellular caspase-8 (FLICE)-like inhibitory protein (cFLIP) was originally identified as an inhibitor of cell-surface death-receptor signaling induced by tumor necrosis factor (TNF), CD95 ligand (CD95 L; also known as FASL), and TNF-related apoptosis-inducing ligand (TRAIL).62 Two predominant splice variants of the FLIP gene, the long form (cFLIPL) and the shorter form (cFLIPS), have been identified. Both forms are capable of protecting cells from apoptosis through competition with procaspase-8 for the recruitment to FAS-associated death domain (FADD), thereby preventing initiator caspase-8 activation.62 In addition, both cFLIP and viral FLIP orthologs (vFLIP) have a prominent role in potently inducing NFκB signaling through the engagement of the IKKγ regulatory subunit of the IκB kinase (IKK) complex.63, 64 These observations seem to indicate that various primary tumor cells and tumors with resistance to death ligands, such as TRAIL, will benefit from enhanced expression of the antiapoptotic and proinflammatory FLIP protein.65
Recent studies by Lee et al.61 have revealed a second important function of both c-FLIP and v-FLIP as effective regulators of autophagy. FLIP depletion by siRNA exaggerates the effect of rapamycin and results in autophagic cell death, whereas the overexpression of FLIP inhibits both starvation and rapamycin-induced autophagy.61 The mechanism of inhibition was found to involve the altering of Atg3 and LC3 interactions during autophagosome elongation. The presence of FLIP competes with LC3 for Atg3 binding and thereby perturbs the lipidation of LC3, but this can be alleviated under stress conditions. As with Bcl-2, this action of FLIP might help maintain the level of autophagy within a physiological range compatible with cell survival and clearance of defective organelles. Interestingly, they identified a mutant FLIP that is defective in both NFκB activation and apoptosis inhibition but remains capable of blocking autophagy as efficiently as wild-type FLIP, suggesting that the antiautophagic role of FLIP is genetically separable from its previously identified antiapoptotic and NFκB activation functions. Thus, it is conceived that autophagy inhibition by FLIP represents a ‘direct hit’ on essential autophagy proteins such as Atg3, rather than an ‘indirect output’ by way of interfering with other signaling pathways.61
Calpain-mediated Atg5 cleavage: two birds, one stone
Post-translational modification of Atg5, a key autophagy effector protein, provides additional insight into the molecular mechanisms of how autophagy is negatively regulated under normal nutritional conditions and in response to stress stimuli.19 Atg5 forms a high-order protein conjugation complex with Atg12 and Atg16, which is essential for autophagosome membrane elongation. Surprisingly, Yousefi et al.19 found that Atg5 also sensitizes tumor cells to various apoptotic stimuli that are not due to enhanced autophagy. In an effort to understand how Atg5 exerts this effect, Yousefi et al.19 identified a cleaved form of Atg5 produced from full-length Atg5 by calpain, a family of Ca2+-dependent non-lysosomal cysteine proteases, in cells exposed to apoptotic stimuli. Atg5 cleavage obliterates its autophagy activity. Instead, the truncated Atg5 translocates to the mitochondria, wherein it associates with Bcl-xL and somehow induces apoptosis. This mechanism has a dual effect in signaling: it enables the direct attenuation of the autophagy response by downregulating Atg5–Atg12 conjugation, while generating unique signals that propagate to mitochondria for the induction of apoptosis. On the basis of this observation, Atg5 cleavage seems to function as a critical switch from protective autophagy to cell death in the presence of apoptotic stimuli. Congruous results were recently provided by Xia et al.66 further demonstrating that calpain1-mediated cleavage of Atg5 can modulate autophagy by adjusting the level of Atg5–Atg12 conjugation in normal cells. Besides Atg5, Beclin1 has also been found to be cleaved by caspases during apoptosis, blunting the autophagy process to promote the apoptotic response.20, 21 Taken together, these findings indicate a critical role for autophagy proteins Atg5 and Beclin1 in the crosstalk between autophagy and apoptosis, while adding calpain and caspases to the growing list of negative regulators of the autophagy track.
Rubicon: gatekeeper of autolysosomal traffic
The autophagosome ‘matures’ through multiple transient interactions with endosomal compartments and lysosomes, to form a hybrid-like organelle called the autolysosome.
The primary function of the autolysosome is to degrade autophagocytosed materials. Along this line, maturation can be an inherently dangerous business, as any misdelivery of cargo to the lysosome can lead to improper proteolytic activation. Currently, little is known about what ‘prepares’ an autophagosome for fusion with a lysosome and what relevant ‘signals’ are involved. However, recent studies point to an important negative regulatory mechanism in autophagosome maturation that is mediated by Rubicon (RUN domain and cysteine-rich domain containing, Beclin1-interacting protein). Rubicon, along with Atg14, was independently identified by two laboratories46, 49 as a novel Beclin1 interactor. Although Atg14 seems conserved from yeast to humans, no yeast counterpart for Rubicon has been found.67 Furthermore, unlike Atg14, which promotes the early stage of autophagosome biogenesis by forming a complex with Beclin1 and PI3KC3, Rubicon negatively regulates a downstream stage of autophagosome maturation by joining the UVRAG, Beclin1, and PI3KC3 complex (Figure 1). Overexpression of Rubicon results in the accumulation of p62, an autophagy cargo, whereas Rubicon depletion induces p62 degradation, suggesting that its overall function is in the inhibition of autophagy.49 In addition, both studies report that the acidification of LC3-II-associated vesicles is severely impaired upon the overexpression of Rubicon,46, 49 indicating that Rubicon expression may restrict autophagic flux to lysosomes. It is currently not clear how Rubicon affects autophagy maturation and its role in diseases remains to be explored.
Although Rubicon is largely considered to be an autophagy-related protein, it is not exclusively present in autophagosomes. Rather, a large portion of Rubicon is present in endosomal compartments.46, 49 Consistent with this observation, Matsunaga et al.46 found that Rubicon also has a role in endocytic trafficking. Overexpression of Rubicon causes defects in endocytic trafficking, whereas the knockdown of Rubicon is associated with enhanced trafficking and degradation of epidermal growth factor receptor (EGFR). Given the previous finding that UVRAG facilitates lysosomal-directed membrane dynamics,68 one could speculate that Rubicon potentially restricts the endocytic forward trafficking to the lysosome by antagonizing UVRAG. Alternatively or additionally, Rubicon may work in concert with UVRAG through different mechanisms to maintain a net balance of overall autophagic flow. Thus, Rubicon may constitute an important component in both autophagic and endocytic trafficking.
Amino acids: negative feedback loop of autophagy
Autophagy is a unique process that degrades long-lived proteins, as well as protein aggregates, to replenish the intracellular amino acid pool, especially in nutrient-deprived cells.23 Indeed, ablation of essential autophagy genes, such as Atg7, significantly dampens the level of intracellular amino acids, as is phenocopied in cells treated with protein synthesis inhibitors.69 Conversely, increased intracellular free amino acids produced during autophagic degradation can reactivate the mTORC1 signaling and thus downregulate autophagy, serving as a self-limiting feedback loop in autophagy regulation.70, 71 Using the Xenopus laevis oocyte as a model, it has been shown that mTORC1 can sense small increases in intracellular amino acids such as leucine, which leads to increased phosphorylation of both p70S6 kinase and its downstream target, ribosomal protein S6.72 Notably, such effects of amino acids on mTORC1 can be independent of stimulation by insulin or other growth factors, but are dependent on autophagic proteolysis.73 A recent study by Yu et al.71 also showed that mTOR signaling is reactivated by prolonged starvation, which attenuates autophagy and restores lysosome homeostasis. This negative feedback loop is envisioned to be part of a homeostatic mechanism required to prevent prolonged or overactivation of autophagy.
The ‘Yin-Yang’ face of p53 in autophagy
The tumor suppressor p53 is often considered the cellular gatekeeper because of its role in sensing and synchronizing numerous stress conditions with cell cycle arrest, DNA repair, senescence, cellular metabolism, apoptosis, and autophagy, in both a transactivation-dependent and/or -independent manner.74 Given the views of autophagy as a cellular stress response, it is unsurprising that several studies have shown that p53 can directly activate autophagy.75, 76, 77 The exact mechanism by which p53 promotes autophagy is not clear, but relates to the ability of p53 to transactivate a group of critical factors that arrest mTOR-mediated anabolic metabolism, including IGF-BP3, PTEN, TSC2, AMPKβ1/β2, and sestrins1/2 (Figure 2).74, 75, 76 Each of these attenuates mTOR signaling and therefore counteracts its repression of autophagy. Damage-regulated autophagy modulator (DRAM) is a p53 transcriptional target and has also been found to positively regulate autophagy.78 Along this line, any impairment of p53 function leads to deregulation of the autophagy signaling pathway and may confer tumorigenesis.
The ‘Yin-Yang’ face of p53 in autophagy regulation. Nuclear p53 positively regulates autophagy in stressed cells through transactivation of target genes. By contrast, cytoplasmic p53 negatively regulates basal autophagy in unstressed cell. Question mark denotes which mechanism of action is unclear. 1, autophagosome initiation; 2, autophagosome membrane elongation; 3, autophagosome–lysosome fusion; 4, degradation and recycling; φ, autophagosome
However, p53's role as an autophagy regulator remains enigmatic, because recent studies by Tasdemir et al. have shown that, in addition to promoting autophagy, p53 also inhibits this process.79 Depletion or pharmacological inhibition of basal levels of p53 in various cell lines, as well as in vivo in mice and Caenorhabditis elegans, was found to induce autophagy, and this increased autophagy conferred resistance to metabolic stress in p53-deficient cells. The mechanism by which p53 suppresses autophagy is not clear, but it seems independent of its transactivation activities.79 Further molecular dissection of p53 by Tasdemir et al. revealed that it is the cytoplasmic p53, not nuclear p53, that mediates suppression of autophagy, reflecting the spatial control of p53 signaling (Figure 2). At first glance, the antiautophagic effects of cytoplasmic p53 cannot be easily reconciled with the view of autophagy as a tumor suppressor mechanism. Yet, many tumor-derived, cytoplasm-restricted forms of p53 are highly oncogenic, which may be partially due to their negative regulation of autophagy, at least in certain contexts.80 Thus, the regulation of autophagy by p53 is multifaceted: stress-induced activation of p53 promotes autophagy, whereas physiological levels of p53 repress autophagy. The dual effects of p53 reflect the coordinated regulation of cellular metabolism, including autophagy, which can be subverted (blunt the positive role (‘yang’) of nuclear p53) or capitalized on (amplify the negative role (‘yin’) of cytoplamsic p53) in malignant transformation.
Conclusions
The pervasiveness and strength of autophagy necessitates the complexity of its negative regulation. Over the past 10 years, an extensive array of negative regulators that limit the amplitude and duration of the autophagic response to confine it to a prescribed range has been identified.1, 2, 3 Defects in these endogenous autophagy inhibitory processes can cause cell toxicity, tissue injury, and predispose the host to many atrophic diseases.12 Conversely, insufficient activation of autophagy causes the accumulation of undesirable components, which can fuel inflammation, trigger necrosis, and induce genomic instability, features underlying the hallmarks of cancer, neurodegeneration, and aging.1, 6, 8 Thus, it is important to envisage autophagy as a tightly regulated dynamic, not only for autophagy per se, but also in the context of its broad connection to other signaling or metabolic pathways. Learning to manipulate this regulatory process of autophagy in the right contexts represents an exciting new challenge. It is likely to generate important advances in our understanding of how autophagy is integrated at a broader level into the cellular network of signaling circuits and how this affects a range of cognitive processes, and is also critical for devising successful therapies for autophagy-associated diseases.
Abbreviations
- TOR:
-
target of rapamycin
- PI3KC3:
-
class III phosphoinositide (PI) 3kinase
- PE:
-
phosphatidylethanolamine
- MEFs:
-
mouse embryonic fibroblasts
- mTOR:
-
mammalian TOR
- PIKK:
-
phosphoinositide 3-kinase-related kinase
- mTORC1:
-
mTOR complex 1
- mTORC2:
-
mTOR complex 2
- Raptor:
-
regulatory-associated protein of mTOR
- LST8:
-
lethal with SEC13 protein 8
- PRAS40:
-
proline-rich AKT-substrate
- Rictor:
-
rapamycin-insensitive companion of mTOR
- SIN1:
-
stress-activated-protein-kinase interacting protein 1
- eIF4E:
-
eukaryotic translation initiation factor 4E
- 4E-BP:
-
eIF4Ebidning protein
- AMPK:
-
AMP-activated protein kinase
- AMPKβ1/β2:
-
the β subunits of AMPK
- PTEN:
-
phosphatase and tensin homolog
- TSC:
-
tuberous sclerosis complex
- PtdIns(3)P:
-
phosphatidylinositol-3-phosphate
- NAF-1:
-
nutrient-deprivation autophagy factor-1
- MTM:
-
myotubularin
- PTP:
-
protein tyrosine phosphatase
- CNM:
-
centronuclear myopathy
- Rubicon, RUN domain and cysteine-rich domain containing:
-
Beclin1 interacting protein
- DAP:
-
death-associated protein
- DAPk:
-
DAP-kinase
- IP3:
-
inositol 1,4,5-triphosphate
- IP3R:
-
IP3 receptor
- Cdks:
-
cycline-dependent kinases
- cFLIP:
-
cellular FLICE-like inhibitory protein
- TNF:
-
tumor-necrosis factor
- TRAIL:
-
TNF-related apoptosis-inducing ligand
- FADD:
-
FAS-associated death domain
- IKK:
-
IκB kinase
- EGFR:
-
epidermal growth factor receptor
- IGF-BP3:
-
insulin-like growth factor binding protein 3
- DRAM:
-
damage-regulated autophagy modulator
References
Levine B, Kroemer G . Autophagy in the pathogenesis of disease. Cell 2008; 132: 27–42.
Yang Z, Klionsky DJ . An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol 2009; 335: 1–32.
Xie Z, Klionsky DJ . Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007; 9: 1102–1109.
Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009; 461: 654–658.
Klionsky DJ, Lane JD . Alternative macroautophagy. Autophagy 2010; 6: 201.
Deretic V . Autophagy in infection. Curr Opin Cell Biol 2010; 22: 252–262.
Liang C, Jung JU . Autophagy genes as tumor suppressors. Curr Opin Cell Biol 2010; 22: 226–233.
Levine B, Kroemer G . Autophagy in aging, disease and death: the true identity of a cell death impostor. Cell Death Differ 2009; 16: 1–2.
Levine B, Yuan J . Autophagy in cell death: an innocent convict? J Clin Invest 2005; 115: 2679–2688.
Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res 2007; 100: 914–922.
De Meyer GR, Martinet W . Autophagy in the cardiovascular system. Biochim Biophys Acta 2009; 1793: 1485–1495.
Sandri M . Autophagy in health and disease: 3. autophagy involvement in muscle atrophy. Am J Physiol Cell Physiol 2010; 298: C1291–C1297.
Cherra SJ, Chu CT . Autophagy in neuroprotection and neurodegeneration: a question of balance. Future Neurol 2008; 3: 309–323.
Virgin HW, Levine B . Autophagy genes in immunity. Nat Immunol 2009; 10: 461–470.
Jackson WT, Giddings Jr TH, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol 2005; 3: e156.
Levine B, Sodora DL . HIV and CXCR4 in a kiss of autophagic death. J Clin Invest 2006; 116: 2078–2080.
Jung CH, Ro SH, Cao J, Otto NM, Kim DH . mTOR regulation of autophagy. FEBS Lett 2010; 584: 1287–1295.
Codogno P, Meijer AJ . Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ 2005; 12 (Suppl 2): 1509–1518.
Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 2006; 8: 1124–1132.
Djavaheri-Mergny M, Maiuri MC, Kroemer G . Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 2010; 29: 1717–1719.
Luo S, Rubinsztein DC . Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ 2010; 17: 268–277.
Heymann D . Autophagy: a protective mechanism in response to stress and inflammation. Curr Opin Investig Drugs 2006; 7: 443–450.
Neufeld TP . TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol 2010; 22: 157–168.
Bjornsti MA, Houghton PJ . The TOR pathway: a target for cancer therapy. Nat Rev Cancer 2004; 4: 335–348.
Huang J, Manning BD . The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J 2008; 412: 179–190.
Yang Z, Klionsky DJ . Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 2010; 22: 124–131.
Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y . Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 2000; 150: 1507–1513.
Chan EY, Longatti A, McKnight NC, Tooze SA . Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13independent mechanism. Mol Cell Biol 2009; 29: 157–171.
Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 2008; 181: 497–510.
Chang YY, Juhasz G, Goraksha-Hicks P, Arsham AM, Mallin DR, Muller LK et al. Nutrient-dependent regulation of autophagy through the target of rapamycin pathway. Biochem Soc Trans 2009; 37: 232–236.
Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 2008; 30: 214–226.
Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 2009; 20: 1981–1991.
Shaw RJ, Cantley LC . Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 2006; 441: 424–430.
Sabatini DM . mTOR and cancer: insights into a complex relationship. Nat Rev Cancer 2006; 6: 729–734.
Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182: 685–701.
Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010; 141: 656–667.
Vergne I, Deretic V . The role of PI3P phosphatases in the regulation of autophagy. FEBS Lett 2010; 584: 1313–1318.
Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122: 927–939.
Long DX, Chang PA, Liang YJ, Yang L, Wu YJ . Degradation of neuropathy target esterase by the macroautophagic lysosomal pathway. Life Sci 2009; 84: 89–96.
Clague MJ, Lorenzo O . The myotubularin family of lipid phosphatases. Traffic 2005; 6: 1063–1069.
Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T et al. Control of autophagy initiation by phosphoinositide 3-phosphatase jumpy. Embo J 2009; 28: 2244–2258.
Shen J, Yu WM, Brotto M, Scherman JA, Guo C, Stoddard C et al. Deficiency of MIP/MTMR14 phosphatase induces a muscle disorder by disrupting Ca(2+) homeostasis. Nat Cell Biol 2009; 11: 769–776.
Laporte J, Bedez F, Bolino A, Mandel JL . Myotubularins, a large disease-associated family of cooperating catalytically active and inactive phosphoinositides phosphatases. Hum Mol Genet 2003; 12 Spec No. 2: R285292.
Tosch V, Rohde HM, Tronchere H, Zanoteli E, Monroy N, Kretz C et al. A novel PtdIns3P and PtdIns(3,5)P2 phosphatase with an inactivating variant in centronuclear myopathy. Hum Mol Genet 2006; 15: 3098–3106.
Erlich S, Mizrachy L, Segev O, Lindenboim L, Zmira O, Adi-Harel S et al. Differential interactions between Beclin 1 and Bcl-2 family members. Autophagy 2007; 3: 561–568.
Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N et al. Two Beclin 1-binding proteins, Atg14 L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009; 11: 385–396.
Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 2006; 8: 688–699.
Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 2007; 9: 1142–1151.
Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT et al. Distinct regulation of autophagic activity by Atg14 L and Rubicon associated with Beclin 1phosphatidylinositol-3-kinase complex. Nat Cell Biol 2009; 11: 468–476.
Maiuri MC, Criollo A, Kroemer G . Crosstalk between apoptosis and autophagy within the Beclin 1 interactome. Embo J 2010; 29: 515–516.
He C, Levine B . The Beclin 1 interactome. Curr Opin Cell Biol 2010; 22: 140–149.
Levine B, Sinha S, Kroemer G . Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy 2008; 4: 600–606.
Chang NC, Nguyen M, Germain M, Shore GC . Antagonism of Beclin 1dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. Embo J 2010; 29: 606–618.
Chen YF, Kao CH, Chen YT, Wang CH, Wu CY, Tsai CY et al. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev 2009; 23: 1183–1194.
Chen R, Valencia I, Zhong F, McColl KS, Roderick HL, Bootman MD et al. Bcl-2 functionally interacts with inositol 1,4,5-trisphosphate receptors to regulate calcium release from the ER in response to inositol 1,4,5-trisphosphate. J Cell Biol 2004; 166: 193–203.
Criollo A, Maiuri MC, Tasdemir E, Vitale I, Fiebig AA, Andrews D et al. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ 2007; 14: 1029–1039.
Vicencio JM, Ortiz C, Criollo A, Jones AW, Kepp O, Galluzzi L et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ 2009; 16: 1006–1017.
E X, Hwang S, Oh S, Lee JS, Jeong JH, Gwack Y et al. Viral Bcl-2-mediated evasion of autophagy aids chronic infection of gammaherpesvirus 68. PLoS Pathog 2009; 5: e1000609.
Furuya T, Kim M, Lipinski M, Li J, Kim D, Lu T et al. Negative Regulation of Vps34 by Cdk Mediated Phosphorylation. Mol Cell 2010; 38: 500–511.
Khalfan WA, Klionsky DJ . Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae. Curr Opin Cell Biol 2002; 14: 468–475.
Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol 2009; 11: 1355–1362.
Safa AR, Day TW, Wu CH . Cellular FLICE-like inhibitory protein (C-FLIP): a novel target for cancer therapy. Curr Cancer Drug Targets 2008; 8: 37–46.
Golks A, Brenner D, Krammer PH, Lavrik IN . The c-FLIP-NH2 terminus (p22FLIP) induces NF-kappaB activation. J Exp Med 2006; 203: 1295–1305.
Chaudhary PM, Jasmin A, Eby MT, Hood L . Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene 1999; 18: 5738–5746.
Yang JK . FLIP as an anti-cancer therapeutic target. Yonsei Med J 2008; 49: 1927.
Xia HG, Zhang L, Chen G, Zhang T, Liu J, Jin M et al. Control of basal autophagy by calpain1 mediated cleavage of ATG5. Autophagy 2010; 6: 61–66.
Zhong Y, Wang QJ, Yue Z . Atg14 L and Rubicon: yin and yang of Beclin 1mediated autophagy control. Autophagy 2009; 5: 890–891.
Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 2008; 10: 776–787.
Onodera J, Ohsumi Y . Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem 2005; 280: 31582–31586.
Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N . Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab 2009; 296: E592602.
Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 2010; 465: 942–946.
Christie GR, Hajduch E, Hundal HS, Proud CG, Taylor PM . Intracellular sensing of amino acids in Xenopus laevis oocytes stimulates p70 S6 kinase in a target of rapamycin-dependent manner. J Biol Chem 2002; 277: 9952–9957.
Beugnet A, Tee AR, Taylor PM, Proud CG . Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem J 2003; 372: 555–566.
Vousden KH, Ryan KM . p53 and metabolism. Nat Rev Cancer 2009; 9: 691–700.
Feng Z, Zhang H, Levine AJ, Jin S . The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA 2005; 102: 8204–8209.
Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G . Autophagy regulation by p53. Curr Opin Cell Biol 2010; 22: 181–185.
Feng Z . p53 Regulation of the IGF-1/AKT/mTOR Pathways and the Endosomal Compartment. Cold Spring Harb Perspect Biol 2010; 2: a001057.
Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006; 126: 121–134.
Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 2008; 10: 676–687.
Morselli E, Tasdemir E, Maiuri MC, Galluzzi L, Kepp O, Criollo A et al. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle 2008; 7: 3056–3061.
Acknowledgements
This work was partly supported by United States Public Health Service Grants CA140964, AI083841, the Leukemia & Lymphoma Society, the Wright Foundation, and the Baxter Foundation (C Liang). We thank Stacy Lee for her critical reading of the paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by D Klionsky
Rights and permissions
About this article
This article is cited by
-
Contribution of endoplasmic reticulum stress, MAPK and PI3K/Akt pathways to the apoptotic death induced by a penicillin derivative in melanoma cells
Apoptosis (2022)
-
DJ-1 binds to Rubicon to Impair LC-3 Associated Phagocytosis
Cell Death & Differentiation (2022)
-
Effects of combination antiretroviral drugs (cART) on hippocampal neuroplasticity in female mice
Journal of NeuroVirology (2021)
-
Cysteinyl leukotriene receptor 1 modulates autophagic activity in retinal pigment epithelial cells
Scientific Reports (2020)
-
Negative regulators of cell death pathways in cancer: perspective on biomarkers and targeted therapies
Apoptosis (2018)