Abstract
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a promising cancer therapeutic agent with cancer-selective apoptogenic activity. It evokes the canonical caspase-mediated cell death pathway through death-inducing signaling complex (DISC) formation. We identified that Peroxiredoxin 6 (Prx6) interacts with caspase-10 and caspase-8 via the death effector domain (DED). Prx6 suppresses TRAIL-mediated cell death in human cancer cells, but not that induced by intrinsic apoptosis inducers such as etoposide, staurosporine, or A23187. Among Prx1–6 members, only Prx6 binds to DED caspases and is most effective in suppressing TRAIL or DED caspase-induced cell death. The antiapoptotic activity of Prx6 against TRAIL is not likely associated with its peroxidase activity but is associated with its ability to bind to DED caspases. Increased expression of Prx6 enhances the binding of Prx6 to caspase-10 but reduces TRAIL-induced DISC formation and subsequently caspase activation. Interestingly, Prx6 is highly upregulated in metastatic gastric cancer cells, which are relatively resistant to TRAIL as compared with primary cancer cells. Downregulation of Prx6 sensitizes the metastatic cancer cells to TRAIL-induced cell death. Taken together, these results suggest that Prx6 modulates TRAIL signaling as a negative regulator of caspase-8 and caspase-10 in DISC formation of TRAIL-resistant metastatic cancer cells.
Similar content being viewed by others
Main
Diverse cellular stresses, such as chemotherapeutic drugs and radiation, disrupt mitochondrial homeostasis and release cytochrome c, leading to apoptosome formation and caspase-9 activation to trigger the intrinsic pathway.1 In the extrinsic pathway, stimulation of cognate death receptors by extracellular death ligands, such as tumor necrosis factor family (TNF) members, recruits the adaptor protein Fas-associated protein with death domain (FADD).2 FADD, in turn, recruits two specific initiator caspases, death effector domain (DED)-containing caspase-8 and caspase-10, through homophilic DED–DED interaction, leading to death-inducing signaling complex (DISC) formation. The DISC facilitates the cleavage and enzymatic activation of DED caspases and downstream executor caspases, caspase-3 and caspase-7.3
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), one of the TNF family members, is the most promising target for the selective eradication of cancer cells without causing toxicity to normal cells.4 Genetic lesions in the components of TRAIL pathway have been found in human tumor samples, implying that TRAIL functions in the surveillance and elimination of developing tumors.5, 6 In addition to DISC formation, caspase-8 is also implicated as an essential component of cytosolic protein complexes containing FADD.7 Further, caspase-8 serves a non-apoptotic role during embryonic development and in the activities of living cells, including cell-cycle progression, T-cell homeostasis, and NF-κB activation.8, 9, 10 Especially, the pivotal roles of caspase-8 and caspase-10 in immunodeficiency11, 12 and tumor development13, 14 highlight the importance of these caspases in the development of disease.
Peroxiredoxin (Prx) is an antioxidant peroxidase enzyme responsible for elimination of hydrogen peroxide, peroxynitrite, and organic hydroperoxides.15 Prxs function in cellular protection against oxidative stress, regulation of cell proliferation, and modulation of intracellular signaling pathways that use hydrogen peroxide as a secondary signal transmitter. The six mammalian isoforms of Prx are produced at a high level and are differentially localized in the cytoplasm, mitochondria, nucleus, and peroxisomes. They are divided into two main subgroups, 1-Cys and 2-Cys Prx, based on the number of conserved catalytic cysteine (peroxidatic cysteine). Prx6 is the only member of 1-Cys Prx and has non-selenium glutathione peroxidase and phospholipase A2 activities.16 Upregulation of Prx6 suppresses intracellular enzyme inactivation, membrane phospholipid peroxidation, and cell death mediated by oxidative stress.17, 18 In addition, Prx6-deficient mice are susceptible to oxidative stress by hypoxia or paraquat treatment, leading to increase of mortality and lung injury.19, 20
Identification of new proteins capable of modulating the activity of DED-harboring caspase is required for a better understanding of diverse cell death pathways and DED-associated human diseases including tumor. Thus, to isolate DED-binding partners, we performed a yeast two-hybrid assay and identified Prx6 as a negative regulator of DED caspases.
Results
Prx6 binds to caspase-10 and caspase-8 through the DED
Prx6 was identified as a caspase-10-binding protein by a yeast two-hybrid screening assay using the DED (DED: amino acids, 1–219) of caspase-10 as bait. To confirm that Prx6 indeed interacts with caspase-10, we performed an in vitro binding assay using the purified GST-fused DED of caspase-10 protein (Figure 1a). Incubation of GST-fused DED with Prx showed that Prx6 binds to caspase-10, while Prx1 fails to do so. FADD was used as a positive control as it is well known to recruit caspase-10 though DED–DED interaction. We explored the binding regions of caspase-10 using HA-fused full-length caspase-10 (amino acids, 1–521) and its serial deletion mutants, including caspase-10DED (1–219), caspase-10ΔDED (220–521), and caspase-10p20 (220–415) (Figure 1b). From immunoprecipitation assays, we found that caspase-10 and caspase-10DED, but not caspase-10ΔDED and caspase-10p20, could interact with Prx6, indicating that DED is responsible for the interaction.
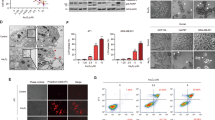
Prx6 binds to caspase-10 and caspase-8 in vitro and in vivo. (a) In vitro binding assay. In vitro translated and 35S-methionine-labeled Prx6, Prx1, or FADD protein was incubated with the purified GST or GST-DED (death effector domain of caspase-10) protein immobilized on glutathione-Sepharose 4B beads. After being pulled down, the bound proteins were separated by SDS-PAGE and detected by autoradiography (upper panel). GST and GST-DED proteins were visualized by coomassie blue staining (lower panel). (b) Intracellular interaction of Prx6 with caspase-10 through DED. HeLa cells were transfected with pHA-Caspase-10 (Casp10), pHA-Caspase-10-DED (Casp10DED), pHA-Caspase-10-ΔDED (Casp10ΔDED), or pHA-Caspase-10-p20 (Casp10p20) for 48 h and cell extracts were subjected to immunoprecipitation (IP) with mouse pre-immune serum (Pre) or anti-HA antibody. The immunoprecipitates were then analyzed by western blotting using anti-Prx6 (upper panel) and anti-HA antibodies (lower panel). Whole cell lysates (WCL) are indicated. Heavy chains (**) of immunoglobulins served as a loading control. (c) Binding of Prx6 to caspase-10 or caspase-8, but not to caspase-9, FADD, or cFLIPL. In vitro translated and 35S-methionine-labeled caspase-10, caspase-9, caspase-8, FADD, or cFLIPL protein was incubated with purified GST, GST-Prx6, GST-FADD, GST-CARD (caspase recruitment domain of Apaf-1), or GST-DED protein. The binding assay was performed as described in a. (d, e) Endogenous interaction between Prx6 and caspase-10 or caspase-8. HeLa cell extracts were immunoprecipitated with rabbit pre-immune serum (Pre) or anti-Prx6 antibody (d) and mouse pre-immune serum (Pre), anti-caspase-10, anti-caspase-9, or anti-caspase-8 antibody (e). The immunoprecipitates were then analyzed by western blotting using anti-Prx6 and the indicated anti-caspase antibodies. Asterisks indicate light chains (*) and heavy chains (**) of immunoglobulins. (f) Visualization of the interaction of Prx6 with caspase-10 or caspase-8 in living cells using BiFC analysis. HeLa cells were co-transfected with equimolar amounts of pPrx6-VN and pCaspase-10-VC, pPrx6-VN and pCaspase-9-VC, pPrx6-VN and pCaspase-8-VC, pbJun-VN and pbFos-VC (a positive control), or pBiFC-VN and pBiFC-VC (a negative control). Green fluorescence images of the complementation were acquired under fluorescence microscope (GFP channel; upper panel) and then overlapped with nuclei Hoechst 33342 staining (Hoechst, middle panel; and Overlay, lower panel)
The binding specificity of Prx6 to DED was then addressed. Like caspase-10, caspase-8 also has DED for the homotypic association with adaptor proteins, whereas caspase-9 has a caspase recruitment domain (CARD). In vitro binding assays showed that GST-fused Prx6 binds to caspase-8 but not to caspase-9, while GST-fused CARD of Apaf-1 binds to caspase-9 (Figure 1c). Prx6 failed to bind to other DED-containing proteins, such as FADD and cFLIPL, besides DED caspases. Cellular interactions of endogenous Prx6 with DED caspases were also observed as assessed with immunoprecipitation assays (Figure 1d and e). The specificity of the anti-Prx6 antibody we generated was validated by western blotting, showing no cross-reactivity with other members of the Prx family (Figure 2e). These results indicate that Prx6 binds to caspase-10 and caspase-8 through DED in vitro and in cells.
Prx6 suppresses cell death mediated by caspase-10 and caspase-8. (a) Inhibition of caspase-10 and caspase-8-induced cell death by Prx6. HeLa cells were co-transfected with pCaspase-12, pCaspase-10, pCaspase-9, or pCaspase-8 together with either pEGFP or pPrx6-GFP in the presence or absence of caspase inhibitor (zVAD; 50 μM zVAD-fmk) for 20 h. Cell death was then assessed. (b, c) Inhibitory kinetics of caspase-10 and caspase-8-induced cell death by Prx6. HeLa cells were transfected with pCaspase-10-GFP (b) or pCaspase-8-GFP (c) and the increasing amounts of pHA-Prx6 for the indicated times, after which cell death was examined. (d, e) Effects of Prx family members on caspase-10 or caspase-8-induced cell death. HeLa cells were co-transfected with pCaspase-10 or pCaspase-8 and either pEGFP (Ctrl), pPrx1-GFP, pPrx2-GFP, pPrx3-GFP, pPrx4-GFP, pPrx5-GFP, or pPrx6-GFP for 20 h, and then cell death was evaluated (d). The expression level of Prx-GFP fusion proteins was examined by western blotting using anti-GFP (upper panel) and anti-Prx6 antibodies (lower panel) (e). (f, g) Antiapoptotic function of wild-type, C47S and C91S Prx6 mutants. HeLa cells were co-transfected with pCaspase-10 or pCaspase-8 and with pEGFP (Ctrl), pPrx6-GFP, pPrx6-C47S-GFP, or pPrx6-C91S-GFP for 20 h, after which cell death was assessed (f). The expression of Prx6-GFP proteins was detected by western blotting using anti-GFP antibody (g). Cell death in a–d, and f was assessed by counting the number of GFP-positive cells showing condensed and fragmented nuclei after staining with EtHD (ethidium homodimer) as described in Materials and Methods. Bars represent mean±S.D. from at least three independent experiments. P values were calculated using t-test and were versus control (#P<0.001; *P<0.01; **P<0.05)
Further, we investigated the interaction of Prx6 with caspase-10 or caspase-8 in living cells using bimolecular fluorescence complementation (BiFC) assay, which enables us to visualize the formation of protein complexes and the location of protein interaction in living cells by fluorescence resonance energy transfer.21 Prx6 was fused to the N-terminal fragment of Venus (VN) (pPrx6-VN) and caspase-10, caspase-9, and caspase-8 were fused to the C-terminal fragment of Venus (VC) (pCaspase-10-VC, pCaspase-9-VC, and pCaspase-8-VC, respectively). Co-expression of pCapase-10-VC or pCaspase-8-VC with pPrx6-VN exhibited fluorescence complementation, which was observed as small green dots predominantly in the cytosol, while co-expression of pCaspase-9-VC with pPrx6-VN showed no green fluorescence (Figure 1f, GFP channel). The fluorescence complementation between the bZIP domains of Jun and Fos (bJun and bFos) fused to VN and VC (bJun-VN and bFos-VC, respectively) was used as a positive control and observed in nucleoli.21 No fluorescence was detected in the reactions containing pPrx6-VN, pCaspase-10-VC, pCaspase-9-VC, or pCaspase-8-VC alone and expression of all of the fusion proteins was verified by western blotting (data not shown).
Prx6 suppresses cell death mediated by caspase-10 and caspase-8
To gain insight into why Prx6 interacts with caspase-10 and caspase-8, we tested whether Prx6 regulates the cell death induced by these caspases. Ectopic expression of Prx6 suppressed cell death induced by caspase-10 or caspase-8, not by caspase-12 or caspase-9, while cell death induced by each of those caspases was blocked by a pan-caspase inhibitor, zVAD-fmk (Figure 2a). Dose-dependent protective activity of Prx6 was clearly observed in cells expressing caspase-10 or caspase-8 and thus undergoing apoptosis (Figure 2b and c).
Among six mammalian isoforms of Prx, we then assessed the selective effect of Prx6 on cell death triggered by caspase-10 or caspase-8. In spite of the protective activities of Prx members harboring peroxidase activity, which reduces the intracellular level of ROS, other Prx members (Prx1–5) except Prx6 were not effective in suppressing cell death induced by caspase-10 or caspase-8 (Figure 2d) and did not bind to the DED of caspase-10 or caspase-8 in vitro (Supplementary Figure 1). The relative expression levels of GFP-fused Prx proteins were confirmed by western blotting (Figure 2e). In addition, Prx6 was most effective in suppressing TRAIL-induced cell death among Prx members (Supplementary Figure 2a). Further, C47S and C91S Prx6 mutants, which contain replaced serine in the peroxidatic cysteine (C47) and the other conserved cysteine (C91), respectively, retained much of their ability to suppress DED caspase-induced cell death, although peroxidase activity-dead C47S Prx6 mutant17 was less effective than wild-type or C91S Prx6 mutant (Figure 2f and g). The C47S and C91S Prx6 mutants also suppressed TRAIL-induced apoptosis (Supplementary Figure 2b). These observations suggest that the antiapoptotic activity of Prx6 in TRAIL signaling may not require its peroxidase activity.
Prx6 inhibits TRAIL-induced cell death of cancer cells
We established a mixed cell population (HeLa/HA-Prx6 Mix) or single-cell clones (HeLa/HA-Prx6 #7 and #12) that conditionally and stably express HA-tagged Prx6 in HeLa cells. Expression of HA-Prx6 in HeLa/HA-Prx6 #7 and #12 stable cells was induced upon the removal of doxycycline to a similar level with endogenous Prx6 (Figure 3a and b). HA-Prx6 was not detected in cells grown in the presence of doxycycline (Figure 3a). As shown in Figure 3c and d, the increased expression of Prx6 significantly suppressed apoptosis induced by TNF family member, TRAIL (55% to ∼18% at 6 h in HeLa/HA-Prx6 #7 and #12 cells) or TNF-α (30% to ∼15% at 6 h in HeLa/HA-Prx6 #7 and #12 cells). However, Prx6 had no effect on apoptosis induced by A23187, etoposide, or staurosporine (Figure 3e–g). Consistent with these results, Prx6 inhibited cell death triggered by the expression of TNF receptor1 (TNFR1) or TRAIL receptors, DR4 and DR5, by about 30%, but not that triggered by truncated Bid (tBid) (Figure 3h). Protein and mRNA levels of receptor signaling proteins in apoptosis were not affected by the induction of Prx6 expression (Figure 3a and b).
Prx6 suppresses cell death triggered by TRAIL or TNF-α, but not by etoposide or staurosporine. (a, b) Generation of HeLa/HA-Prx6 stable cell lines using pEBNA-1 (Tet-Off). HeLa cells were transfected with pYR-HA control vector (HeLa/Hph) or pYR-HA-Prx6 (HeLa/HA-Prx6 Mix) and selected with 250 μg/ml hygromycin for 2 weeks to generate mixed population. Single clones (HeLa/HA-Prx6 #7 and #12) were then isolated from HeLa/HA-Prx6 Mix cells. HeLa/Hph and HeLa/HA-Prx6 stable cells were grown in the presence (+) or absence (−) of 1 μg/ml doxycycline (Dox) for 48 h, after which cell extracts were prepared and the levels of proteins and mRNAs involved in apoptosis pathway were examined by western blotting (a) and RT-PCR analysis (b), respectively. (c–g) Inhibition of TRAIL or TNF-α-induced cell death by Prx6. HeLa/Hph and HeLa/HA-Prx6 cell lines were maintained in the presence or absence of 1 μg/ml doxycyline for 2 days, and then left untreated (Mock) or exposed to 100 ng/ml TRAIL (c), 30 ng/ml TNF-α and 1 μg/ml cycloheximide (TNF-α; d), 2 μM A23187 (e), 50 μM etoposide (f), or 0.2 μM staurosporine (g) for the indicated times. Cell death was quantified using LIVE/DEAD Viability/Cytotoxicity kit. (h) Prx6 suppresses cell death induced by ectopic expression of TRAIL and TNF receptors, but not that by the truncated Bid (tBid). HeLa/Hph and HeLa/HA-Prx6 #7 cells were grown in doxycycline-free media for 48 h and co-transfected with pEGFP (Ctrl) or with pDR4, pDR5, pTNFR1, or ptBid for 20 h. Cell death was then assessed as described in Figure 2a. Values in c–h are mean±S.D. from four independent experiments and P values were calculated using t-test (*P<0.01; **P<0.05)
Conversely, when we reduced the expression of Prx6 using Prx6 short hairpin RNAs (Prx6 shRNA #1 or #2), we found that HeLa cells became significantly sensitive to cell death mediated by caspase-10 or caspase-8, but not to that mediated by caspase-12 or caspase-9 (Figure 4a and b). Further, Prx6 shRNAs also sensitized HeLa cells to cell death induced by TRAIL (25%) (Figure 4c) or TNFα (21%) (Figure 4d), but did not affect cell death by A23187 (Figure 4e) or staurosporine (data not shown). Knockdown of Prx6 itself did not trigger cell death in the absence of cell-death stimuli (data not shown). These results suggest that Prx6 has an essential role in the receptor-mediated apoptosis, which is coordinated with caspase-10 and caspase-8.
Downregulation of Prx6 expression sensitizes cancer cells to TRAIL-mediated cell death. (a, b) Depletion of Prx6 sensitizes HeLa cells to caspases-10 and caspase-8. HeLa cells were transfected with control pshRNA (Ctrl) or one of two different pPrx6 shRNA #1 and #2 for 20 h and western blotting was performed with anti-Prx6, anti-Prx1, and anti-α-tubulin antibodies (a). HeLa cells were co-transfected for 20 h with pshRNA, pHA-Prx6, pPrx6 shRNA #1, or pPrx6 shRNA #2, and either pEGFP or pCaspase-GFP as indicated. After staining with EtHD, cell death was quantified by counting the number of both GFP and EtHD-positive cells from total GFP-positive cells (b). (c–e) Knockdown of Prx6 sensitizes HeLa cells to TRAIL or TNFα-induced cell death. HeLa cells were co-transfected with pEGFP and either pshRNA or pPrx6 shRNAs for 20 h and then left untreated (Mock) or treated with 100 ng/ml TRAIL (c), 30 ng/ml TNF-α and 1 μg/ml cycloheximide (TNF-α; d), or 2 μM A23187 (e) for the indicated times. Cell death was quantified as described in b. The results in b–e represent mean±S.D. of three independent experiments
Prx6 inhibits TRAIL-induced activation of caspase-10 and caspase-8 by interfering with death-inducing signaling complex (DISC) formation
We explored whether Prx6 directly modulates enzymatic activation of caspase-10 and caspase-8 to inhibit TRAIL-induced cell death. From western blotting, we found that compared with HeLa/HA-Prx6 #7 control cells, the proteolytic processing of caspase-10 and caspase-8 was apparently reduced by the expression of Prx6 in HeLa/HA-Prx6 #7 stable cells after removal of doxycycline (Figure 5a). The processed forms of downstream caspase-3 and PARP were also less detected in HeLa/HA-Prx6 #7 stable cells expressing Prx6 than in control cells. Consistently, enzymatic activation of caspase-10 and caspase-8 as well as caspase-3 was significantly suppressed at early time points ranging from 0.5 to 1.5 h after treatment with TRAIL (Figure 5b–e). These results suggest that Prx6 functions as an apoptosis suppressor that directly inhibits the activation of caspase-10 and caspase-8 in TRAIL-mediated apoptosis.
Prx6 inhibits enzymatic activation of caspases-10 and caspase-8 during TRAIL signaling. (a) Blockade of caspase processing by Prx6 during TRAIL-induced apoptosis. HeLa/HA-Prx6 #7 cells were incubated in the presence or absence of doxycycline for 48 h and then exposed to 100 ng/ml TRAIL for the indicated times. Cell extracts were examined by western blotting using the indicated antibodies. (b–e) Suppression of TRAIL-induced enzymatic activation of caspases by Prx6. HeLa/HA-Prx6 #7 cells were incubated in the presence or absence of doxycyline for 2 days and then treated with 100 ng/ml TRAIL for the indicated times. Cell extracts were prepared and examined for caspase activity using 50 μM fluorogenic caspase substrates, AEVD-AFC (b), IETD-AFC (c), DEVD-AFC (d), and LEHD-AMC (e). Bars represent mean±S.D. (n=3)
The observation that Prx6 interacts with caspase-10 and caspase-8 and regulates TRAIL-induced cell death led us to examine the effects of Prx6 on the formation of DISC. Immunoprecipitation assay showed that after exposure to TRAIL, caspase-10, caspase-8, and FADD were detected in DR4- or DR5-containing DISC, which was isolated from HeLa/HA-Prx6 #7 cells cultured in the presence of doxycycline (Figure 6a). However, caspase-10 and caspase-8 were not or were much less detected in the DISC isolated from TRAIL-treated HeLa/HA-Prx6 #7 cells expressing HA-Prx6, while the recruitment of FADD to DISC was not affected. Furthermore, endogenous interaction of Prx6 with caspase-10 or caspase-8, which was observed in HeLa/HA-Prx6 #7 stable cells grown in the presence of doxycycline, was inhibited upon treatment with TRAIL. More interestingly, these interactions still remained in HeLa/HA-Prx6 #7 cells after withdrawal of doxycycline (Figure 6a). FADD was not detected in the immunoprecipitates containing Prx6 independently of TRAIL treatment. These results suggest that Prx6 may inhibit the formation of DISC in the cells exposed to TRAIL through its interaction with DED caspases.
Increased expression of Prx6 suppresses the binding of caspase-10 to FADD for DISC formation during TRAIL signaling. (a) Lack of caspase-10 and caspase-8 in DR4 or DR5-containing DISC complex in HeLa/HA-Prx6 #7 cells exposed to TRAIL. HeLa/HA-Prx6 #7 cells were grown in the presence or absence of doxycyline for 2 days and then left untreated or exposed to 100 ng/ml TRAIL for 15 min. Cells were lysed and subjected to immunoprecipitation (IP) assay with anti-DR4 and anti-DR5 antibodies together or with anti-Prx6 antibody alone. The immunoprecipitates were examined by western blotting using the indicated antibodies. Asterisks (**) indicate heavy chains of antibodies. (b) Competitive binding of DED caspases to Prx6 or FADD. In vitro translated and 35S-methionine-labeled caspase-10 or caspase-8 was incubated with 5 μg GST-Prx6 protein alone or together with the increasing amounts of purified His-FADD protein. Pull-down assays were performed using glutathione-Sepharose 4B beads as described in Figure 1a. After SDS-PAGE separation, Prx6-bound caspase-10 and caspase-8 were visualized by autoradiography (upper panel) and GST-Prx6 protein was shown by Coomassie blue staining (lower panel)
During DISC formation of apoptosis, FADD is known to interact with DED caspases through DED, whereas Prx6 is also believed to bind to DED caspases in live cells. Thus, we further assessed a possible mechanism for the regulation of protein–protein interaction between Prx6, FADD, and DED caspases. From in vitro binding assays, we found that the interaction of Prx6 with caspase-10 or caspase-8 was gradually decreased by the presence of increasing amounts of FADD protein in the reaction (Figure 6b). The disruption of Prx6–caspase-8 interaction by FADD seems to be more dramatic than that of Prx6–caspase-10. These results imply that there is competitive binding of DED caspases to Prx6 and FADD and thus, the relative amounts of DED caspase-binding partners may be important to regulate the recruitment of DED caspase into DISC during TRAIL signaling.
Upregulation of Prx6 expression in metastatic gastric cancer cells renders TRAIL resistance to cancer cells
Consistent with the previous studies in breast cancers,22, 23 we also found that the expression of Prx6 was significantly elevated in highly metastatic gastric cancer cells, including SNU-5, SNU-16, SNU-216, SNU-601, and SNU-668,24 as compared with primary cancer cells (Figure 7a). These TRAIL-resistant cancer cells showed no significant change in the level of cFLIP, a known suppressor of TRAIL cytotoxicity in metastatic cancers25 (Figure 7a). Further, we observed that those metastatic gastric cancer cells were relatively resistant to TRAIL-induced cell death (Figure 7b), indicating that there may be a correlation between the level of Prx6 expression and TRAIL resistance in metastatic gastric cancer cells. Then, silencing of Prx6 expression in SNU-216 gastric cancer cells by using two independent Prx6 shRNAs increases cells’ susceptibility to TRAIL-induced cell death (Figure 7c). Thus, these results indicate that upregulation of Prx6 expression in metastatic gastric cancer cells decreases cells’ susceptibility to TRAIL-induced cell death and thereby may contribute to tumor surveillance in TRAIL-resistant metastatic cancers.
Upregulation of Prx6 protein in metastatic gastric cancer cells confers tolerance to TRAIL-induced cell death. (a) Upregulation of Prx6 protein in metastatic gastric cancer cells. The lysates of primary and metastatic gastric cancer cells were examined by western blotting using anti-Prx6, anti-caspase-10, anti-caspase-8, anti-cFLIPs, and anti-β-actin antibodies (upper panel). Prx6 signals were quantified by densitometry using the histogram feature in Adobe Photoshop (lower panel). (b) Resistance of metastatic gastric cancer cells to TRAIL-induced cell death. The gastric cancer cells were untreated (Mock) or treated with 100 ng/ml TRAIL for 6 h, after which cell death was measured using LIVE/DEAD Viability/Cytotoxicity kit. (c) Sensitization of TRAIL-resistant metastatic cancer cells to TRAIL by downregulation of Prx6 expression. SNU-216 cells were co-transfected with pEGFP and either pshRNA or pPrx6 shRNAs (#1 and #2) for 36 h, and then treated with 100 ng/ml TRAIL for 8 h. Cell death was quantified by EtHD staining. The results represent mean±S.D. (n=4). Cell extracts were examined by western blotting using anti-Prx6 and anti-β-actin antibodies to show the depletion of Prx6 protein (right panel)
Discussion
DED is a prototypical protein interaction domain that functions predominantly in the regulation and execution of programmed cell death and additionally in the control of cell proliferation. In most cases, DED-containing proteins form a protein complex through homotypic DED interactions. However, non-homotypic interaction of DED-containing protein has also been reported. DED-lacking DED-associated factor (DEDAF) and caspase-8 and caspase-10-associated RING protein (CARP) interact with caspase-10 and caspase-8 in the cytosol and regulate the function of DED caspases.26, 27 Similarly, DED-lacking Prx6 interacts with DED-containing caspase-10 and caspase-8 but not with structurally similar CARD-containing caspase-9. Any structural motifs of Prx6 responsible for these interactions are not clarified yet.
Despite the fact that Prx1–6 belong to the peroxiredoxin family harboring peroxidase activity, it is clear that only Prx6 can interact with DED caspases and concomitantly suppress cell death mediated by DED caspases. The amino-acid sequence identity of Prx6, the only member of the 1-Cys subgroup, with other Prxs is low (<20%), whereas that among the 2-Cys subgroup members is high (60–80%). The low level of homology of Prx6 with other Prxs might confer DED caspase-binding specificity to Prx6. Several previous studies have demonstrated that Prx6 has unique binding partners such as surfactant protein A and saitohin.28, 29 Interestingly, only Prx6 uses glutathione as a physiological reductant, while other Prxs use thioredoxin.16
Then, how does Prx6 regulate TRAIL-mediated cell death through DED caspases? The antiapoptotic activity of Prx6 can be exerted mainly by two possible mechanisms. One is that Prx6 suppresses cell death triggered by TRAIL through its peroxidase activity. However, other Prxs (Prx1–5) do not affect much TRAIL-induced cell death, and peroxidase activity-dead C47S Prx6 mutant, which does not show structural change, still has antiapoptotic activity, indicating that the peroxidase activity of Prx6 is not sufficient to regulate TRAIL or DED caspase-mediated cell death. The other, major mechanism is that Prx6 binds to and sequestrates DED caspases that are not to be recruited into the DISC complex even in the presence of TRAIL signaling. This is the reason why Prx6 effectively suppresses DED caspase and TRAIL-induced cell death and fails to inhibit cell death induced by the intrinsic cell death stimuli and caspase-9. In addition, of six Prx members, only Prx6 has acidic Ca2+-independent phospholipase A2 activity provided by the catalytic activity of Ser32 (S32). We observed that phospholipase activity-dead S32A Prx6 mutant considerably lost its antiapoptotic activity. As this mutant shows a marked change of secondary structure and protein folding,30 the role of phospholipase activity of Prx6 in TRAIL-induced cell death is not clear yet.
Under normal physiological conditions Prx6 interacts with the proform of caspase-10 and caspase-8 through DED, but not with active caspases. It is also notable that the interaction of caspase-10 and caspase-8 with Prx6 seems to be weaker than that with FADD, implying that the binding affinity of Prx6 to DED caspase is lesser than that of FADD. Thus, it is plausible that upon stimulation with TRAIL, DED caspase dissociates from Prx6 and is recruited into DISC following the high binding affinity to FADD. Previous studies demonstrated that TRAIL treatment triggers the production of reactive oxygen species (ROS), resulting in intracellular oxidizing conditions.31 Interestingly, we observed that in vitro binding of Prx6 to caspase-10 was decreased by increased ROS level with the treatment of hydrogen peroxide, but was increased by decreased ROS level with the treatment of ROS scavenger, dithiothreiton (DTT) (Supplementary Figure 3). Thus, we propose a hypothesis that cytosolic accumulation of ROS also may reduce the binding affinity of Prx6 to DED caspase and enhance the chances of FADD to interact with DED caspases to form DISC.
TRAIL receptors are defined as metastasis suppressor proteins that reduce the metastatic propensity of tumor cells.32, 33 Inactivation of TRAIL signaling by mutation of TRAIL receptors, DR4 and DR5, was specifically observed in metastatic cancer, and downregulation of TRAIL receptors promotes metastasis of cancer cells.34, 35 Also, metastatic oral cancer cells showed greater TRAIL resistance than their primary oral tumors.36 Recently, caspase-8 has been identified as a metastasis suppressor because of its function as a downstream mediator of TRAIL-mediated apoptosis.37 Suppression of caspase-8 expression occurs during the establishment of cancer metastases. Interestingly, the expression of Prx6 is significantly elevated in highly metastatic breast cancer cells as compared with its parental counterparts,23 and overexpression of Prx6 leads to a more invasive phenotype and metastatic capability.22 We also found the overexpression of Prx6 in metastatic gastric cancer cells. Thus, we propose that the elevated levels of Prx6 in cancers may interfere with the dissociation of DED caspase from Prx6, making DED caspase to be hardly recruited into DISC and causing greater resistance to TRAIL in metastatic cancer cells. These events will confer a metastatic phenotype to TRAIL-resistant primary cancer cells during early metastasis formation.
Taken together, our observation that Prx6 regulates TRAIL-induced cell death by binding to and modulating the activation of DED caspases may provide a molecular basis for the role of Prx6 as a negative regulator of metastasis suppressor.
Materials and Methods
Yeast two-hybrid screening assay
The DED region (amino acids 1–219) of caspase-10 was cloned into pLexA vector (Clontech, Mountain View, CA, USA) and used as bait for conventional yeast two-hybrid assay. Jurkat cDNA library cloned into pB42AD vector (Agilent Technologies, Stratagene Products Division, La Jolla, CA, USA) was screened as described.38
Construction of plasmids
Human Prx6 cDNA was subcloned into pcDNA3-HA (Invitrogen, Carlsbad, CA, USA), pEGFP (Clontech), and pYR3323-HA vector to generate HA or GFP fusion protein (pHA-Prx6, pPrx6-GFP, and pYR-HA-Prx6, respectively). Human caspase-10 cDNA was subcloned into pcDNA3-HA (pHA-Caspase-10) or pEGFP-N1 vector (pCaspase-10-GFP). To construct serial deletion mutants of caspase-10, caspase-10-DED (amino acids 1–219), caspase-10-ΔDED (220–521), and caspase-10-p20 (220–415) were amplified by PCR and subcloned into pcDNA3-HA vector (pHA-Caspase-10-DED, pHA-Caspase-10-ΔDED, and pHA-Caspase-10-p20). Human Prx1–5 cDNAs were subcloned into pEGFP to generate GFP fusion proteins (pPrx1-GFP, pPrx2-GFP, pPrx3-GFP, pPrx4-GFP, and pPrx5-GFP). All Prx constructs were verified by DNA sequencing analysis. Prx6 was fused to N-terminal Venus fragments of pBiFC-VN vector (pPrx6-VN) and caspase-10, caspase-9, and caspase-8 were fused to C-terminal Venus fragments of pBiFC-VC vector (pCaspase-10-VC, pCaspase-9-VC, and pCaspase-8-VC).21
BiFC assay
BiFC assay was performed as described.21 HeLa cells were transfected with VN or VC-fusion construct alone or in combination and incubated at 37°C for 20 h. After staining cell nuclei with Hoechst 33342, fluorophore formation in the living cells was imaged using a fluorescence microscope (Olympus, Hamburg, Germany).
Mutagenesis and shRNA
Point mutants of Prx6 (Prx6 (C47S) and Prx6 (C91S)) were generated by a Quickchange Site-Directed Mutagenesis kit (Stratagene) using synthetic oligonucleotides containing mutations in the corresponding positions. All mutants were verified by DNA sequencing analysis. To construct pPrx6 shRNA, heteroduplex oligomers containing gene-specific sequences (pPrx6 shRNA #1 (188-AGAGGAATGTTAAGTTGAT-206) and pPrx6 shRNA #2 (176-CAGAATTTGCCAAGAGGAA-194)) were synthesized, annealed, and cloned into the BglII and HindIII sites of pSUPER.neo vector (pshRNA) (OligoEngine, Seattle, WA, USA).
Cell culture and stable cell lines
HeLa and HeLa EBNA cells39 (HeLa Tet-Off (Clontech)-derived cells stably expressing EBNA-1) were cultured in Dulbecco’s modified Eagle's medium (Invitrogen) with 10% (v/v) fetal bovine serum (Invitrogen). HeLa EBNA cells were transfected with an episomal expression vector pYR-HA or pYR-HA-Prx6 that contains the tetracycline-regulated promoter for regulating the expression of an inserted gene, oriP, for episome replication, and the selection marker for hygromycin B. The transfected cells were cultivated in a medium supplemented with 250 μg/ml hygromycin B (Clontech) and 1 μg/ml doxycycline (Sigma-Aldrich, St Louis, MO, USA) for 2 weeks to generate a control cell line (HeLa/Hph) and a mixed population cell line (HeLa/HA-Prx6 Mix). A single cell was cultivated to form stable cell clones (HeLa/HA-Prx6 #7 and #12) and the expression levels were examined with western blotting. Stable cell clones were maintained in doxycycline-deficient medium for 2 days to induce the expression of HA-tagged Prx6 and employed for further experiments. Human gastric cancer cell lines (SNU-1, -5, -16, -216, -484, -601, -668, and -719) were obtained from the Korean Cell Line Bank (Seoul, Korea) and maintained in RPMI1640 medium (HyClone Laboratories, Logan, UT, USA) with 10% (v/v) fetal bovine serum.
Transfection, cell death assessment, and viability assay
Expression plasmids were transfected into 1 × 105 cells per well in six-well dishes using LipofectAMINE Reagent (Invitrogen) following the manufacturer's instructions. Cell death was assessed by counting the number of GFP and EtHD-positive cells after staining the cells with 0.5 mM ethidium homodimer (EtHD; Molecular Probes, Eugene, OR, USA) at 37°C for 15 min. Cell death was examined using LIVE/DEAD Viability/Cytotoxicity kit (Molecular Probes).
Antibodies and western blotting
Polyclonal rabbit anti-Prx6 antibody was generated by immunizing rabbits with purified GST-Prx6 protein, following standard immunization procedures. The following antibodies were used for western blot analyses: caspase-10 (MBL, M059-3), caspase-9 (Cell Signaling, #9502), caspase-8,40 cleaved caspase-3 (Cell Signaling, #9661), DR4 (Abcam, Cambridge, MA, USA, 13890), DR5 (Abcam, 47179), PARP-1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA, sc-8007), FADD (BD Pharmingen, San Diego, CA, USA, 610400), α-tubulin (Sigma, T5168), β-actin (Sigma, A2668), and GFP (Santa Cruz Biotechnology, sc-8334) antibodies. Cells were lysed in ice-cold RIPA buffer (50 mM Tris-Cl (pH 8.0), 15 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin A) and sonicated briefly. Cell lysates were clarified by centrifugation, resolved by SDS-PAGE, and transferred onto the PVDF membrane (Millipore, Billerica, MA, USA). Immunoblots were visualized by enhanced chemiluminescence method.
In vitro binding assay
GST-fusion proteins cloned into pGEX vector (GE Healthcare, Amersham Biosciences, Uppsala, Sweden) (GST-Prx6, GST-Caspase-10-DED (amino acids 1–219), GST-Caspase-8-DED (1–197), GST-FADD, and GST-CARD (CARD of Apaf-1; 1–601)) were expressed and purified using glutathione-Sepharose 4B (Amersham Biosciences). The purified GST-fusion proteins were incubated with 35S-methionine-labeled proteins, which were translated in vitro using a TNT-coupled transcription/translation system (Promega, Madison, WI, USA) in ice-cold binding buffer (50 mM Tris-Cl (pH 7.4), 150 mM NaCl, 0.1% NP-40, and protease inhibitors) at 4°C for 3 h with gentle rocking. After the beads were washed five times with 1 ml binding buffer, bound proteins were eluted in 2 × SDS loading buffer, separated by SDS-PAGE, and visualized by autoradiography.
Immunoprecipitation
To detect the endogenous interaction of Prx6 and either caspase-10 or caspase-8, HeLa cells were lysed in 1 ml RIPA buffer containing protease inhibitor cocktail. After the lysates were centrifugated at 15 000 × g for 30 min and pre-cleared with 20 μl protein G-Sepharose at 4°C for 1 h, the resulting supernatant was incubated with pre-immune serum or the appropriate antibody at 4°C for 2 h. After adding 50 μl of protein G-Sepharose or protein A-Sepharose (Amersham Biosciences), the mixtures were incubated at 4°C for an additional 3 h. The immunoprecipitates were washed five times with RIPA buffer and analyzed by western blotting.
RT (reverse transcription)-PCR
Cells were lysed and RNA was extracted using Trizol (Invitrogen), according to the manufacturer's protocol. PCR was performed for 20 cycles using the following primer sets: Prx6 (5′-ATGCCCGGAGGTCTGCTTCTCGGGGACGTG-3′, 5′-TTAAGGCTGGGGTGTGTAGCGGAGGTATTT-3′), caspase-10 (5′-ATGAAATCTCAAGGTCAACATTGGTATT-3′, 5′-ACCTTGTTTCTCTAGAAATGCCAGGAAA-3′), caspase-9 (5′-ATGGACGAAGCGGATCGGCGGCTCCTGC-3′, 5′-GTCCACTGGTCTGGGTGTTTCCGGTCTG-3′), caspase-8 (5′-ATGGACTTCAGCAGAAATCTTTATGATA-3′, 5′-GGCAGAAATTTGAGCCCTGCCTGGTGTC-3′), caspase-3 (5′-ATGGAGAACACTGAAAACTCAGTGGATT-3′, 5′-TGCTGTGGAGTATGCATACAAGAAGTCG-3′), FADD (5′-ATGGACCCGTTCCTGGTGCTG-3′, 5′-GGACGCTTCGGAGGTAGATGC-3′), DR5 (5′-ATGGAACAACGGGGACAG-3′, 5′-TTAGGACATGGCAGAGTC-3′), Apaf-1 (5′-ATGGATGCAAAAGCTCGAAATTGTTTGCTT-3′, 5′-ACTCTCATCCTGATCCAACCGTGTGCAAAG-3′), and β-actin (5′-GAGCTGCCTGACGGCCAGG-3′, 5′-CATCTGCTGGAAGGTGGAC-3′).
Caspase activity assay
Cells treated with TRAIL were homogenized in cold lysis buffer (20 mM Hepes (pH 7.6), 0.1% Chaps, 100 mM KCl, 2 mM DTT, 0.5 mM Na-EDTA, 50 μg/ml calpeptin, and protease inhibitors) and clarified by centrifugation at 18 000 × g for 1 h. The supernatant (40 μg of protein) was incubated in reaction buffer (20 mM Hepes (pH 7.6), 0.1% Chaps, 2 mM DTT, and 1% sucrose) with 50 μM fluorogenic caspase substrates, DEVD-AFC, IETD-AFC (BD Pharmingen), LEHD-AMC, or AEVD-AFC, at 37°C for 1 h. Fluorescence (excitation, 405 nm; emission, 505 nm and 535 nm) was measured using a fluorescence microplate reader.
Statistical analysis
Data are expressed as mean±S.D. Statistical comparisons between groups were performed using one-way analysis of variance (ANOVA) followed by Bonferroni's test or by the Student's t-test as appropriate. Probabilities of P<0.05 were considered statistically significant.
Abbreviations
- DED:
-
death effector domain
- DISC:
-
death-inducing signaling complex
- Prx:
-
peroxiredoxin
- TRAIL:
-
tumor necrosis factor-related apoptosis-inducing ligand
- BiFC:
-
bimolecular fluorescence complementation
References
Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 1997; 91: 479–489.
Thomas LR, Henson A, Reed JC, Salsbury FR, Thorburn A . Direct binding of Fas-associated death domain (FADD) to the tumor necrosis factor-related apoptosis-inducing ligand receptor DR5 is regulated by the death effector domain of FADD. J Biol Chem 2004; 279: 32780–32785.
Sprick MR, Rieser E, Stahl H, Grosse-Wilde A, Weigand MA, Walczak H . Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but can not functionally substitute caspase-8. EMBO J 2002; 21: 4520–4530.
Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med 1999; 5: 157–163.
Finnberg N, Klein-Szanto AJ, El-Deiry WS . TRAIL-R deficiency in mice promotes susceptibility to chronic inflammation and tumorigenesis. J Clin Invest 2008; 118: 111–123.
Takeda K, Hayakawa Y, Smyth MJ, Kayagaki N, Yamaguchi N, Kakuta S et al. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat Med 2001; 7: 94–100.
Wang L, Du F, Wang X . TNF-alpha induces two distinct caspase-8 activation pathways. Cell 2008; 133: 693–703.
Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL et al. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 1998; 9: 267–276.
Olson NE, Graves JD, Shu GL, Ryan EJ, Clark EA . Caspase activity is required for stimulated B lymphocytes to enter the cell cycle. J Immunol 2003; 170: 6065–6072.
Salmena L, Lemmers B, Hakem A, Matysiak-Zablocki E, Murakami K, Au PY et al. Essential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunity. Genes Dev 2003; 17: 883–895.
Wang J, Zheng L, Lobito A, Chan FK, Dale J, Sneller M et al. Inherited human Caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell 1999; 98: 47–58.
Chun HJ, Zheng L, Ahmad M, Wang J, Speirs CK, Siegel RM et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature 2002; 419: 395–399.
Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA et al. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med 2000; 6: 529–535.
Shin MS, Kim HS, Kang CS, Park WS, Kim SY, Lee SN et al. Inactivating mutations of CASP10 gene in non-Hodgkin lymphomas. Blood 2002; 99: 4094–4099.
Rhee SG, Chae HZ, Kim K . Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med 2005; 38: 1543–1552.
Chen JW, Dodia C, Feinstein SI, Jain MK, Fisher AB . 1-Cys peroxiredoxin, a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. J Biol Chem 2000; 275: 28421–28427.
Kang SW, Baines IC, Rhee SG . Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J Biol Chem 1998; 273: 6303–6311.
Manevich Y, Sweitzer T, Pak JH, Feinstein SI, Muzykantov V, Fisher AB . 1-Cys peroxiredoxin overexpression protects cells against phospholipid peroxidation-mediated membrane damage. Proc Natl Acad Sci USA 2002; 99: 11599–11604.
Wang X, Phelan SA, Forsman-Semb K, Taylor EF, Petros C, Brown A et al. Mice with targeted mutation of peroxiredoxin 6 develop normally but are susceptible to oxidative stress. J Biol Chem 2003; 278: 25179–25190.
Wang Y, Feinstein SI, Manevich Y, Ho YS, Fisher AB . Peroxiredoxin 6 gene-targeted mice show increased lung injury with paraquat-induced oxidative stress. Antioxid Redox Signal 2006; 8: 229–237.
Hu CD, Chinenov Y, Kerppola TK . Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 2002; 9: 789–798.
Chang XZ, Li DQ, Hou YF, Wu J, Lu JS, Di GH et al. Identification of the functional role of peroxiredoxin 6 in the progression of breast cancer. Breast Cancer Res 2007; 9: R76.
Li DQ, Wang L, Fei F, Hou YF, Luo JM, Zeng R et al. Identification of breast cancer metastasis-associated proteins in an isogenic tumor metastasis model using two-dimensional gel electrophoresis and liquid chromatography-ion trap-mass spectrometry. Proteomics 2006; 6: 3352–3368.
Park JG, Yang HK, Kim WH, Chung JK, Kang MS, Lee JH et al. Establishment and characterization of human gastric carcinoma cell lines. Int J Cancer 1997; 70: 443–449.
Hamai A, Richon C, Meslin F, Faure F, Kauffmann A, Lecluse Y et al. Imatinib enhances human melanoma cell susceptibility to TRAIL-induced cell death: relationship to Bcl-2 family and caspase activation. Oncogene 2006; 25: 7618–7634.
Zheng L, Schickling O, Peter ME, Lenardo MJ . The death effector domain-associated factor plays distinct regulatory roles in the nucleus and cytoplasm. J Biol Chem 2001; 276: 31945–31952.
McDonald III ER, El-Deiry WS . Suppression of caspase-8- and -10-associated RING proteins results in sensitization to death ligands and inhibition of tumor cell growth. Proc Natl Acad Sci USA 2004; 101: 6170–6175.
Wu YZ, Manevich Y, Baldwin JL, Dodia C, Yu K, Feinstein SI et al. Interaction of surfactant protein A with peroxiredoxin 6 regulates phospholipase A2 activity. J Biol Chem 2006; 281: 7515–7525.
Gao L, Tse SW, Conrad C, Andreadis A . Saitohin, which is nested in the tau locus and confers allele-specific susceptibility to several neurodegenerative diseases, interacts with peroxiredoxin 6. J Biol Chem 2005; 280: 39268–39272.
Manevich Y, Reddy KS, Shuvaeva T, Feinstein SI, Fisher AB . Structure and phospholipase function of peroxiredoxin 6: identification of the catalytic triad and its role in phospholipid substrate binding. J Lipid Res 2007; 48: 2306–2318.
Choi K, Ryu SW, Song S, Choi H, Kang SW, Choi C . Caspase-dependent generation of reactive oxygen species in human astrocytoma cells contributes to resistance to TRAIL-mediated apoptosis. Cell Death Differ 2010; 17: 833–845.
Rinker-Schaeffer CW, O’Keefe JP, Welch DR, Theodorescu D . Metastasis suppressor proteins: discovery, molecular mechanisms, and clinical application. Clin Cancer Res 2006; 12: 3882–3889.
Steeg PS . Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer 2003; 3: 55–63.
Shin MS, Kim HS, Lee SH, Park WS, Kim SY, Park JY et al. Mutations of tumor necrosis factor-related apoptosis-inducing ligand receptor 1 (TRAIL-R1) and receptor 2 (TRAIL-R2) genes in metastatic breast cancers. Cancer Res 2001; 61: 4942–4946.
Zhuang L, Lee CS, Scolyer RA, McCarthy SW, Zhang XD, Thompson JF et al. Progression in melanoma is associated with decreased expression of death receptors for tumor necrosis factor-related apoptosis-inducing ligand. Hum Pathol 2006; 37: 1286–1294.
Vigneswaran N, Wu J, Nagaraj N, Adler-Storthz K, Zacharias W . Differential susceptibility of metastatic and primary oral cancer cells to TRAIL-induced apoptosis. Int J Oncol 2005; 26: 103–112.
Stupack DG, Teitz T, Potter MD, Mikolon D, Houghton PJ, Kidd VJ et al. Potentiation of neuroblastoma metastasis by loss of caspase-8. Nature 2006; 439: 95–99.
Chang JW, Choi H, Kim HJ, Jo DG, Jeon YJ, Noh JY et al. Neuronal vulnerability of CLN3 deletion to calcium-induced cytotoxicity is mediated by calsenilin. Hum Mol Genet 2007; 16: 317–326.
Park Y, Hwang YP, Lee JS, Seo SH, Yoon SK, Yoon JB . Proteasomal ATPase-associated factor 1 negatively regulates proteasome activity by interacting with proteasomal ATPases. Mol Cell Biol 2005; 25: 3842–3853.
Kim IK, Chung CW, Woo HN, Hong GS, Nagata S, Jung YK . Reconstitution of caspase-8 sensitizes JB6 cells to TRAIL. Biochem Biophys Res Commun 2000; 277: 311–316.
Acknowledgements
We thank Dr. JB Yoon (Yonsei University, Korea) for HeLa EBNA cells and Dr. TK Kerppola for BiFC plasmids. H Choi was partly supported by the BK2 program. This work was supported by grants from the CRI-Acceleration Research Program, Nuclear Research Grant (BAERI), and Cancer Project (to Y-K Jung) funded by the Ministry of Education, Science and Technology (MEST) and of Human Health and Welfare in Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by J Silke
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Choi, H., Chang, JW. & Jung, YK. Peroxiredoxin 6 interferes with TRAIL-induced death-inducing signaling complex formation by binding to death effector domain caspase. Cell Death Differ 18, 405–414 (2011). https://doi.org/10.1038/cdd.2010.113
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2010.113
Keywords
This article is cited by
-
Exploring the role of PRDX4 in the development of uterine corpus endometrial carcinoma
Medical Oncology (2024)
-
Amelioration of intracellular stress and reduction of neural tube defects in embryos of diabetic mice by phytochemical quercetin
Scientific Reports (2016)
-
miR-24-3p Regulates Progression of Gastric Mucosal Lesions and Suppresses Proliferation and Invasiveness of N87 Via Peroxiredoxin 6
Digestive Diseases and Sciences (2016)
-
Inhibition of transketolase by oxythiamine altered dynamics of protein signals in pancreatic cancer cells
Experimental Hematology & Oncology (2013)
-
Regulation in the targeting of TRAIL receptor 1 to cell surface via GODZ for TRAIL sensitivity in tumor cells
Cell Death & Differentiation (2012)