Abstract
In order to infer population structure at the individual level, we estimated individual inbreeding coefficients and examined the relationship between geographical distance and genetic relatedness from polymorphic microsatellite data for a population of Mimulus guttatus that has an intermediate selfing rate. Expected heterozygosities for five microsatellites ranged from 0.79 to 0.93. The population inbreeding coefficient was calculated to be 0.19 (SE=0.023). A method-of-moments estimator developed by Ritland (1996b) was used to estimate the distribution of inbreeding among and relatedness between individuals of a natural population. The mean individual inbreeding coefficient (F=0.16) did not differ significantly from the population-level estimate. Most of the individuals appeared to be outbred, and there were very few plants that had estimated inbreeding coefficients greater than one-half. Individuals sampled from one transect showed significantly more inbreeding than individuals sampled along the other (P=0.005). There was no apparent relationship between interplant distance (range: 0–14 m) and mean genetic relatedness between individuals. These results represent the first application of polymorphic microsatellites to estimate fine-scale genetic population structure.
Similar content being viewed by others
Introduction
The genetic structure of a population can have profound effects on its evolutionary potential. Through the use of polymorphic genetic markers such as allozymes, there is now a large number of studies that describe population structure in terms such as the average level of inbreeding within a population or the average genetic differentiation between populations or subpopulations (Hamrick & Godt, 1990). In contrast, very few studies use markers to infer population structure at the level of individual organisms. This is unfortunate, because knowledge of the extent to which individuals differ in their inbreeding histories and the degree of genetic relatedness between pairs of individuals is important for many areas of ecology and evolution.
For example, if individuals vary in their inbreeding histories, then we expect statistical associations to exist between diploid genotypes at different loci (Haldane, 1949; Kimura, 1958). Knowing the variance in individual inbreeding coefficients, and therefore the relative magnitude of these associations (referred to as identity disequilibrium), is important for the interpretation of studies of natural selection at individual loci (Ohta & Cockerham, 1974; Charlesworth, 1990; Houle, 1994) and on quantitative traits (Willis, 1996). In addition, the existence of individual variation in inbreeding coefficient implies that traditional quantitative genetic methods for predicting the evolutionary response to selection in outbred (Lande & Arnold, 1983) or fully inbred populations (Mather & Jinks, 1982) are not appropriate. Instead, one can use the distribution of inbreeding histories in a partially inbred population, in conjunction with alternative quantitative genetic techniques, to evaluate evolutionary potential and short-term response to selection (Kelly, 1999a,b). Finally, knowledge of the individual inbreeding coefficients enables the study of many aspects of plant mating system evolution, such as the relationship between inbreeding history and traits like fitness or the probability of self-fertilization (Schultz & Willis, 1996).
Knowing the genetic relatedness between pairs of individuals is useful for analyses of population structure, kin selection and quantitative genetics in the field. For example, the relationship between pairwise relatedness and physical distance can give an indication of the spatial scale at which population differentiation occurs (e.g. Loiselle et al., 1995). Also, the distribution of relatedness in groups can give indications of the potential for kin selection (e.g. Schuster & Mitton, 1991). Finally, the relationship between relatedness and phenotypic similarity of individuals can be used to study quantitative genetic inheritance in nature (Ritland & Ritland, 1996; Ritland, 1996a).
The two primary reasons for the scarcity of studies of population structure at the individual level are the lack of adequate statistical methods and sufficiently informative genetic markers, because typical markers such as allozymes usually exhibit low to moderate levels of polymorphism. Recently, Ritland (1996b) developed a marker-based method for estimating individual inbreeding and pairwise relatedness in natural populations. He pointed out in this paper that his method-of-moments estimator (MME) should be particularly useful for highly polymorphic molecular markers, because standard errors of the estimates decrease as the number of alleles per locus increases. Microsatellites, stretches of DNA consisting of short nucleotide repeats, are ideal molecular markers for such studies because they are codominant and often highly polymorphic within populations. We have recently developed a large number of polymorphic microsatellite markers in the common yellow monkey flower Mimulus guttatus (Kelly & Willis, 1998). In this paper, we use Ritland’s MME in concert with microsatellite data to examine the genetic structure at the level of individuals in a partially self-fertilizing population. The goals of this research are to (i) estimate the distribution of individual inbreeding coefficients for a natural population, and (ii) determine the extent to which relatedness between individuals declines with geographical distance.
Methods
Study species and population
Mimulus guttatus (Scrophulariaceae, 2n=28) is a self-compatible wildflower that lives throughout much of western North America and is common in moist areas such as stream banks, roadsides or wet meadows. Populations of M. guttatus have been reported to vary substantially in their rates of self-fertilization from highly selfing to predominantly outcrossing (Ritland & Ganders, 1987; Ritland, 1990; Willis, 1993b; Awadalla & Ritland, 1997). The flowers of M. guttatus are large and cross-fertilization occurs via bee pollinators.
We studied an annual population of M. guttatus that is located on a seasonally moist slope of Iron Mountain in Oregon’s Western Cascades. Though the population occurs over a small area of localized seepage from melting snow, the population is quite large and consists of tens of thousands of individuals. Seeds germinate shortly after the snow melts in early June and flowering occurs about a month later. Because this area normally receives little rainfall during these months, all plants die by the end of July. The Iron Mountain population has a mixed mating system and estimates of self-fertilization rates over two years range from about 10% to 25% (Willis, 1993b).
Field sampling and DNA isolation
Juvenile M. guttatus plants were collected from the Iron Mountain population in early summer 1997, and then transplanted to the University of Oregon greenhouse. A total of 106 individuals were collected as pairs of neighbouring plants every 0.5 m along two parallel 14 m transects. The two transects were separated by 14 m, and were situated perpendicular to the downhill slope. Plants were collected in this manner in the hope of obtaining a sample maximizing the contrast between genetically similar and dissimilar individuals. Ninety-six plants survived transplantation, and their DNA was extracted from corollas as previously described (Lin & Ritland, 1995). Microsatellite data were obtained for 92 of these plants.
Microsatellite analysis
Preliminary work identified four (AAT)n loci and one (AG)n microsatellite locus as being highly polymorphic in the Iron Mountain population of M. guttatus. Three of the five microsatellites (AAT211, AAT300 and AAT356) have been described previously (Kelly & Willis, 1998). Microsatellite alleles at these five loci were detected following PCR amplification of genomic DNA with radioactive primers (see Table 1 for primer sequences and PCR conditions). The forward primer was labelled in a standard end-labelling reaction using [γ-P32]ATP and T3 polynucleotide kinase reaction (10 μM forward primer, 1× kinase buffer (Promega), 1 U/5 μL kinase (Promega)). PCR reactions were set up in 10 μL volumes in 96-well microtitre plates, each well containing 40 ng of template DNA, 1× thermal reaction buffer (Promega), 1–2 mM MgCl2, 0.2 μM forward and reverse primers, 0.02 mM dNTPs, 0.5 U Taq DNA polymerase (Promega); some reactions were supplemented with an additional 50 mM KCl (Table 1). The samples were overlaid with mineral oil and microsatellite loci were amplified in an MJR thermal cycler. A 1.5-min denaturation step at 94°C was followed by 20 cycles of 40 s of denaturation at 94°C, 1 min of annealing at varying temperatures (generally 4–5°C below Tm, Table 1), and 40 s of extension at 74°C, followed by a final 2 min of extension at 74°C. Samples were then mixed with 5 μL of formamide loading dye and denatured for 2 min at 75°C. From each reaction mixture, 3.5 μL was loaded onto a 6% denaturing acrylamide gel. Finally, gels were dried on Whatman paper, and overlaid with X-ray film. The film was developed following 12–24 h of exposure at 20°C. An allele-sizing ladder was included on the gels at 16-lane intervals. This ladder was a sequencing reaction performed on a previously cloned AAT microsatellite locus for which the repeat region extends through the relevant base pairs sizes. Data were obtained for a total of 92 individuals, although a few individuals had incomplete genotypic data (see Table 2).
Statistical analyses
At each of the five loci, expected heterozygosity (He) and Wright’s population inbreeding coefficient (F) were calculated according to the formulae

respectively, where pi is the observed frequency of the ith allele and Ho is the frequency of heterozygotes observed in the sample population. Additionally, He was calculated separately for each transect. Differences between F-values at each of the five loci were examined via bootstrapping by resampling individual plants with replacement. The bootstrap method was applied to each locus (with 5000 resamplings), as well as to the difference between all pairs of loci (with 5000 resamplings). The standard error of each locus was obtained through bootstrapping. Additionally, differences in F-values were calculated directly between all locus pairs. A standard Bonferroni correction (Rice, 1989) was performed to infer simultaneously one significance level (P=0.05) for comparisons between all loci. A weighted average F was calculated from the single-locus estimates with bootstrapped variances as the weights. Individual inbreeding coefficients were calculated using the method-of-moments estimator (MME) for the two-gene relationship described by Ritland (1996b). A simplified, multilocus estimator for individual levels of inbreeding is given by

where ρ represents the inbreeding coefficient F, Pil is the estimate of the frequency of the ith allele at the lth locus, S indicates whether the locus is homozygous for the ith allele and n represents the number of unique alleles at the locus. If an individual is homozygous for allele i at locus l, Sil=1, otherwise Sil=0.
The frequency distribution of inbreeding coefficients for all individuals was examined, as well as the frequency distribution of only those individuals having genotypes at all five loci. To investigate how estimates of inbreeding coefficients were affected by locus number, two loci were randomly removed from the subset of individuals for which genotypes at all five loci were determined. The mean five-locus individual inbreeding coefficient was compared with the mean of the newly created three-locus inbreeding coefficients and differences were examined by a t-test. Additionally, the correlation between five-locus inbreeding coefficients and three-locus inbreeding coefficients was determined. Finally, a t-test was performed to examine differences between the two transects in the mean inbreeding among individuals (inbreeding values were log transformed for analysis).
Using the same multilocus MME (Ritland, 1996b) given above, values of relatedness between pairs of individuals in the population were calculated. In this case, ρ represents r, the coefficient of kinship, between two individuals. Pil is the estimate of the frequency of the ith allele at the lth locus, Sil is the average over four ways that a pair of identical alleles can be sampled by taking one allele from each individual and n is the number of unique alleles at the locus. The 92 individuals were paired so that relatedness values for all possible combinations of plants were calculated. Distances between individuals were then entered into a separate matrix. Relatedness was plotted as a function of distance and the correlation coefficient was calculated. Comparisons were made to investigate the structure of relatedness within the population. Relatedness among nearest neighbours (pairs from the same point along the transect) was compared to relatedness among all other pairs. Additionally, levels of relatedness within each transect were compared to levels between transects. For each of these comparisons, statistical significance of differences was evaluated using t-tests.
Results
Population-level diversity and inbreeding
Genotypes at microsatellite loci were determined for a total of 92 individuals, though only 49 individuals have genotypic data at all five loci. Genotypic data were obtained for at least three of the five loci for nearly all individuals (six individuals have genotypes scored at fewer than three loci). The five microsatellite loci were extremely variable in this population, with large numbers of alleles per locus and correspondingly high expected heterozygosities ranging from 0.79 to 0.93 (Table 2).
The single-locus estimates of the population-level inbreeding coefficient, F, ranged from 0.09 to 0.29 (Table 2). Nearly all alleles were found in both transects and expected heterozygosities were very similar when calculated separately for each transect (data not shown). Bootstrapping the differences between the distributions of locus pairs found no significant differences in F for all pairs of loci (based on bootstrap confidence intervals, P > 0.05) except AAT9–AG19, and AAT211–AAT356 (based on bootstrap confidence intervals, P=0.01 and P=0.03, respectively). Using the standard Bonferroni technique, a single significance level was simultaneously inferred between all loci comparisons, revealing that F does not significantly differ across the five loci (P > 0.05). The variance-weighted average F across loci is 0.191 with a 95% confidence interval between 0.145 and 0.237 (SE=0.023).
Individual inbreeding coefficients
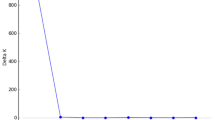
Using the methods-of-moments estimator formulated by Ritland (1996b), inbreeding coefficients were estimated for each individual based on genotypic data from all loci available (Fig. 1a, x¯=0.16, SE=0.025, n=92). Additionally, the frequency distribution of inbreeding values for only those individuals having genotypic data at all five loci was constructed (Fig. 1b, x¯=0.17, SE=0.032, n=49). Large numbers of individuals are completely outbred and there is an exponential decline towards increasing levels of inbreeding. Both of these estimates of individual inbreeding coefficients have means which fall within the 95% confidence interval for the calculated population-level inbreeding coefficient (F=0.145–0.237, P=0.05). The variance in inbreeding coefficients was 0.057 when all data were considered, and 0.051 when data were used only from those individuals scored for all five loci.
To examine the effect of loci number on the individual inbreeding estimates, genotypic data were removed from two randomly chosen loci for the 49 individuals with genotypes at all five loci, and new inbreeding coefficients were scored using the remaining three loci. There was a significant correlation between inbreeding coefficients calculated from five loci and those calculated from three (r=0.632, P < 0.0001). Additionally, the mean with three loci (x¯=0.18, SE=0.041, n=49) did not differ significantly from the mean of the five-locus genotypes (t=−0.479, P=0.634).
The distributions of inbreeding coefficients for individuals within each linear transect were compared. The spatial distribution of inbreeding among all 92 individuals reveals a significantly higher level of inbreeding in the lower transect (Fig. 2; transect 1: x¯=0.21; transect 2: x¯=0.08; t=−2.90, P=0.005).
Genetic relatedness and geographical distance
Using Ritland’s MME, relatedness coefficients were calculated for each possible pair of individuals and plotted as a function of interplant distance. For the M. guttatus individuals sampled in this population, mean relatedness does not vary with distance (multiple R2=0.0002, F=0.89, P=0.34). Mean relatedness between nearest-neighbour pairs was higher than relatedness between pairs considered at increasing interplant distances, though not significantly so (nearest-neighbour pairs: x¯=0.0030; all other pairs: x¯=−0.0056; t=1.14, P=0.25). Levels of relatedness were not significantly different between the two transects (transect 1: x¯=−0.0047; transect 2: x¯=−0.0053; t=0.29, P=0.77). Additionally, relatedness levels calculated for pairs within each transect were similar to those calculated for pairs between different transects (between transects: x¯=−0.0061; t=−0.87, P=0.38; t=−0.45, P=0.65 for comparisons with transects 1 and 2, respectively).
Discussion
Distribution of individual inbreeding coefficients
This study presents the first estimates of individual inbreeding coefficients for a natural population obtained from data on highly variable microsatellites. Inspection of the distribution of individual inbreeding coefficients in the Iron Mountain population of M. guttatus indicates that most individuals are completely outbred, with a sharp decline in the frequency of individuals having increased levels of inbreeding. The mean inbreeding coefficient of 0.16 reflects the fact that the majority of individuals appeared to be only slightly inbred. As discussed below, even neighbouring plants were essentially unrelated. For this reason, it is likely that most of this inbreeding is caused by self-fertilization and not biparental inbreeding. If all inbreeding resulted from selfing, we would expect individual inbreeding coefficients to take discrete values of 0, 0.5, 0.75, and so on. The fact that our estimated inbreeding coefficients often fall between these values is probably a result of sampling error. Very few individuals appear to be the products of more than one generation of self-fertilization. This lack of highly inbred plants probably reflects both the low frequency of such individuals expected with low to moderate selfing rates (see below) and the fact that inbred plants suffer tremendous inbreeding depression in this population (Willis, 1993a,b, 1999). As the plants genotyped in this study were collected as juveniles, a stage in the life cycle before most of the lifetime inbreeding depression is expressed (Willis, 1993a,b, 1999), we expect to observe selfed individuals derived from outcrossed parents but only relatively rarely from inbred parents.
Because individuals differ in their history of inbreeding, we should have observed identity disequilibrium between different loci (Haldane, 1949; Bennett & Binet, 1956; Kimura, 1958). Variation in the history of inbreeding causes outbred individuals to be more heterozygous at all loci and inbred individuals to be more homozygous than expected based on their single-locus genotype frequencies. For example, one should observe an excess in the frequency of two-locus heterozygotes, standardized by the product of the two single-locus expected heterozygosities, that is equal to the variance in individual inbreeding coefficients if the loci are unlinked (Haldane, 1949; Bennett & Binet, 1956; Kimura, 1958). In this study we did find an excess of double homozygotes and heterozygotes. The magnitude of this excess is approximately what one would expect given the distribution of individual inbreeding coefficients. For example, the observed excess of double heterozygotes, averaged over all pair-wise comparisons of the markers, is equal to 0.013, which, given the observed variation among pairs of loci, is approximately what one expects based on the expected heterozygosities and the estimate for variance in individual inbreeding coefficient of about 0.05. In the absence of inbreeding depression, we might have expected this identity disequilibrium to have been even greater.
Interestingly, a spatial division in the distribution of individual inbreeding levels was observed between the two transects within the population. Inbreeding coefficients were significantly higher for individuals in the lower transect than for those plants located in the uphill transect. Given that the marker alleles are distributed evenly throughout both transects and that expected heterozygosities are similar in each, it is likely that the observed difference in inbreeding coefficients among the transects is caused by true variation in rates of self-fertilization and not by artefacts of population structure such as a Wahlund effect resulting from downhill seed dispersal. This would suggest that ecological factors (e.g. plant density) or quantitative traits (e.g. flower size) may differ between the two transects, perhaps affecting pollinator visitation rates.
Given the strong similarity between inbreeding estimates using three loci and those using five, it is likely that the distribution of inbreeding coefficients determined by all individuals (because only six individuals have fewer than three loci) is not biased by the different loci numbers used for different plants. Apparently, accuracy in the methods-of-moments estimator does not significantly decline when only three loci are used rather than all five. Indeed, Ritland’s MME allows estimations of inbreeding coefficients with data from as few as one locus, though a large statistical error is inherent in such a small sample size (Ritland, 1996b).
Population inbreeding coefficient
As expected, the mean individual inbreeding coefficient of 0.16 was not significantly different from the population-level estimate of 0.19. The population inbreeding coefficient estimated in the present study is within the range of values reported from previous studies utilizing allozymes and microsatellites, which have found tremendous variation in inbreeding levels between M. guttatus populations (Ritland & Ganders, 1987; Ritland, 1990; Willis, 1993b; Latta & Ritland, 1994; Awadalla & Ritland, 1997). The population-level inbreeding coefficients at Iron Mountain were estimated in a previous study (Willis, 1993b) as 0.05 (SE=0.16) and 0.06 (SE=0.23) for the years 1989 and 1990, respectively. The value of 0.19 for F in the present study indicates significant inbreeding in the Iron Mountain population, and it is not significantly different from these earlier estimates.
Relatedness between individuals at different spatial scales
No spatial structure of relatedness within the Iron Mountain population of M. guttatus was apparent from analyses conducted in this study. Neither a linear nor a nonlinear relationship was detected between relatedness and physical distance. Though mean relatedness between nearest neighbours was slightly higher than mean relatedness between plants with greater interplant distances, this difference was not significant. These results are somewhat surprising, given the fact that other populations of M. guttatus exhibit spatial structure on a local scale (Ritland & Ganders, 1987; Ritland & Ritland, 1996) and the observation of short flight distances of the Megachilid bees which are the primary visitor in this population (mean flight distance between flowers=30 cm, n=62; G. Smick, unpubl. data).
Mean relatedness between nearest neighbours (interplant distance=0) was quite low (r=0.0003) compared with previous estimates for levels of relatedness between M. guttatus individuals within distances of 1 m (Ritland & Ritland, 1996). Utilizing the methods-of-moments estimator, Ritland & Ritland (1996) calculated mean relatedness between individuals at distances of 1 m as 0.125 and 0.052 for a meadow and stream habitat, respectively. Both previous estimates of relatedness within 1 m were significantly different from 0, as well as from the value of nearest-neighbour mean relatedness calculated in the present study. Ritland & Ritland (1996) believe that the observed discrepancy in levels of relatedness between the two habitats is almost certainly caused by differences in water-mediated seed dispersal. This view suggests that gene flow via seed dispersal is greater in the stream habitat, leading to lower levels of relatedness between individuals separated by small distances (such as 1 m). Similarly, it is possible that seed dispersal is relatively extensive in the Iron Mountain population. As the population occurs along a fairly steep incline, perhaps there is greater opportunity for downhill seed dispersal. Another explanation could be that greater wind-mediated seed dispersal is afforded by the exposed nature of the hillside meadow. Additionally, gene flow by way of pollen transfer could occur over larger scales in the Iron Mountain population as compared to the habitats investigated by Ritland & Ritland (1996). Given the considerable variation between populations of M. guttatus for other genetic parameters such as the outcrossing rate and the average inbreeding coefficient, it is perhaps not surprising that the spatial distribution of relatedness should also vary. Future experiments should examine the extent of gene flow by measuring pollen and seed dispersal.
Biparental inbreeding (mating between close relatives) is thought to be an unavoidable consequence of a population structure in which relatives are clustered together. In one M. guttatus population, Leclerc-Potvin & Ritland (1994) estimated that biparental inbreeding accounted for 20–40% of the total observed self-fertilization (25%). By contrast, because single- and multilocus estimates of outcrossing rates were not significantly different, Willis (1993b) concluded that biparental inbreeding was negligible in the Iron Mountain population. That the present study found little evidence of microspatial relatedness structure supports the claim that biparental inbreeding is infrequent in this population. Moreover, this suggests that observed differences in levels of individual inbreeding result almost entirely from differences in rates of self-fertilization. Overall, these results indicate that interplant distance may be a poor indication of relatedness between individuals in the Iron Mountain population. Similarly, other studies have been unable to demonstrate conclusively that genetic relatedness declines with distance (Waser, 1987; Loiselle et al., 1995; Hamilton, 1997).
References
Awadalla, P. and Ritland, K. (1997). Microsatellite variation and evolution in the Mimulus guttatus species complex with contrasting mating systems. Mol Biol Evol. 14: 1023–1034.
Bennett, J. H. and Binet, F. E. (1956). Association between Mendelian factors with mixed selfing and random mating. Heredity. 10: 51–55.
Charlesworth, D. (1990). The apparent selection on neutral marker loci in partially inbreeding populations. Genet Res. 57: 159–175.
Haldane, J. B. S. (1949). The association of characters as a result of inbreeding and linkage. Ann Eugen. 15: 15–23.
Hamilton, M. B. (1997). Genetic fingerprint-inferred population subdivision and spatial genetic tests for isolation by distance and adaptation in the coastal plant Limonium carolinianum. Evolution. 51: 1457–1468.
Hamrick, J. L. and Godt, M. J. W. (1990). Allozyme diversity in plant species. In: Brown, A. H. D., Clegg, M. T., Kahler, A. L. and Weir, B. S. (eds) Plant Population Genetics, Breeding, and Genetic Resources pp. 43–63. Sinauer Associates, Sunderland, MA.
Houle, D. (1994). Adaptive distance and the genetic basis of heterosis. Evolution. 48: 1410–1417.
Kelly, A. J. and Willis, J. H. (1998). Polymorphic microsatellite loci in Mimulus guttatus and related species. Mol Ecol. 7: 769–774.
Kelly, J. K. (1999a). Response to selection in partially self-fertilizing populations. I. Selection on a single trait. Evolution. 53: 336–349.
Kelly, J. K. (1999b). Response to selection in partially self-fertilizing populations. II. Selection on multiple traits. Evolution. 53: 350–357.
Kimura, M. (1958). Zygotic frequencies in a partially self-fertilizing population. Ann Report Nat Inst Genet, Japan. 8: 104–105.
Lande, R. and Arnold, S. J. (1983). The measurement of selection on correlated characters. Evolution. 37: 1210–1226.
Latta, R. and Ritland, K. (1994). The relationship between inbreeding depression and prior inbreeding among populations of four Mimulus taxa. Evolution. 48: 806–817.
Leclerc-Potvin, C. and Ritland, K. (1994). Modes of self-fertilization in Mimulus guttatus (Scrophulariaceae): a field experiment. Am J Bot. 81: 199–205.
Lin, J. -Z. and Ritland, K. (1995). Flower petals allow simpler and better isolation of DNA for plant RAPD analyses. Plant Mol Biol Rep. 13: 215–218.
Loiselle, B. A., Sork, V. L., Nason, J. and Graham, C. (1995). Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot. 82: 1420–1425.
Mather, K. and Jinks, J. L. (1982). Biometrical Genetics 3rd edn. Chapman & Hall, London.
Ohta, T. and Cockerham, C. C. (1974). Detrimental genes with partial selfing and effects on a neutral locus. Genet Res. 23: 191–200.
Rice, W. R. (1989). Analyzing tables of statistical data. Evolution. 43: 223–225.
Ritland, K. (1990). Inferences about inbreeding depression based upon changes of the inbreeding coefficient. Evolution. 44: 1230–1241.
Ritland, K. (1996). A marker-based method for inferences about quantitative inheritance in natural populations. Evolution. 50: 1062–1073.
Ritland, K. (1996). Estimators for pairwise relatedness and individual inbreeding coefficients. Genet Res. 67: 175–185.
Ritland, K. and Ganders, F. R. (1987). Covariation of selfing rates with parental gene fixation indices within populations of Mimulus guttatus. Evolution. 41: 760–771.
Ritland, K. and Ritland, C. (1996). Inferences about quantitative inheritance based on natural population structure in the yellow monkeyflower, Mimulus guttatus. Evolution. 50: 1074–1082.
Schultz, S. and Willis, J. H. (1996). Individual variation in inbreeding depression: the roles of inbreeding history and mutation. Genetics. 141: 1209–1223.
Schuster, W. S. F. and Mitton, J. B. (1991). Relatedness within clusters of a bird-dispersed pine and the potential for kin interactions. Heredity. 67: 41–48.
Waser, N. M. (1987). Spatial genetic heterogeneity in a population of the montane perennial plant Delphinium nelsonii. Heredity. 58: 249–256.
Willis, J. H. (1993a). Effects of different levels of inbreeding on fitness components in Mimulus guttatus. Evolution. 47: 864–876.
Willis, J. H. (1993b). Partial self-fertilization and inbreeding depression in two populations of Mimulus guttatus. Heredity. 71: 145–154.
Willis, J. H. (1996). Measures of phenotypic selection are biased by partial inbreeding. Evolution. 50: 1501–1511.
Willis, J. H. (1999). The role of genes of large effect on inbreeding depression in Mimulus guttatus. Evolution. in press.
Acknowledgements
We would like to thank John Kelly and Peter J. Russell for helpful comments and discussion, as well as Alan Kelly for invaluable laboratory advice. This research was supported in part by NSF grants DEB-9419884 and DEB-9727578 to J.H.W.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sweigart, A., Karoly, K., Jones, A. et al. The distribution of individual inbreeding coefficients and pairwise relatedness in a population of Mimulus guttatus. Heredity 83, 625–632 (1999). https://doi.org/10.1038/sj.hdy.6886020
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6886020
Keywords
This article is cited by
-
Multi-level patterns of genetic structure and isolation by distance in the widespread plant Mimulus guttatus
Heredity (2020)
-
Population viability analysis predicts decreasing genetic diversity in ex situ populations of the Itasenpara bitterling Acheilognathus longipinnis from the Kiso River, Japan
Ichthyological Research (2017)
-
Decreasing genetic diversity in wild and captive populations of endangered Itasenpara bittering (Acheilognathus longipinnis) in the Himi region, central Japan, and recommendations for conservation
Conservation Genetics (2014)
-
Direct estimation of the mutation rate at dinucleotide microsatellite loci in Arabidopsis thaliana (Brassicaceae)
Heredity (2009)
-
Testing the rare-alleles model of quantitative variation by artificial selection
Genetica (2007)