Abstract
In recent years, the quagga mussel, Dreissena rostriformis bugensis, native to the Dnieper and Bug Limans of the northern Black Sea, has been dispersed by human activities across the basin, throughout much of the Volga River system, and to the Laurentian Great Lakes. We used six published microsatellite markers to survey populations throughout its native and introduced range to identify relationships among potential source populations and introduced ones. Mussels from 12 sites in Eurasia, including the central Caspian Sea and one in North America (Lake Erie), were sampled. Field surveys in the Volga River basin suggested that the species first colonized the middle reach of the river near Kubyshev Reservoir, and thereafter spread both upstream and downstream. Evidence of considerable gene flow among populations was observed and genetic diversity was consistent with a larger, metapopulation that has not experienced bottlenecks or founder effects. We propose that high gene flow, possibly due to multiple invasions, has facilitated establishment of quagga mussel populations in the Volga River system. The Caspian Sea population (D. rostriformis rostriformis (=distincta)) was genetically more distinct than other populations, a finding that may be related to habitat differences. The geographical pattern of genetic divergence is not characteristic of isolation-by-distance but, rather, of long-distance dispersal, most likely mediated by commercial ships' ballast water transfer.
Similar content being viewed by others
Introduction
Nonindigenous species (NIS) have attracted much attention in recent years due to their often-negative impacts on native biodiversity or ecosystem function. The invasion and subsequent rapid expansion into the Laurentian Great Lakes by two dreissenid mussels endemic to the Ponto-Caspian region of Europe is a prime example of such an aquatic invasion (eg Mills et al, 1999; Ricciardi and MacIsaac, 2000). Dreissena polymorpha, the zebra mussel, was discovered in the Great Lakes in 1988, while a second dreissenid, the quagga mussel (see taxonomic discussion below), was discovered in 1992 (Hebert et al, 1989; May and Marsden, 1992). Both species have since spread throughout the lower Great Lakes and the St Lawrence River, although the zebra mussel has achieved a much broader distribution that includes much of temperate eastern North America (Mills et al, 1996). Similarly, in Europe, D. polymorpha dispersed rapidly to new habitats as early as the 18th century, as interbasin connections were established (eg Kinzelbach, 1992; Minchin et al, 2002), while quagga mussels maintained a relatively restricted distribution until recently. It was confined to the Bug Liman, Ukraine, until the 1940s and was not reported from neighboring Black Sea estuaries, reservoirs and canals until the 1990s (eg Kharchenko, 1995; Mills et al, 1996; Table 1). In 1992, quagga mussels were first observed in the Volga River system in Kuybyshev Reservoir and the upper part of Saratov Reservoir (Antonov, 1993; Orlova and Shcherbina, 2002). Range expansion has since been both rapid and extensive. The species range now encompasses a 3000 km stretch of the Volga River system from Uglich and Rybinsk Reservoirs in the north to the Volga River Delta, including the northern Caspian Sea shallows, in the south (Figure 1; Orlova et al, 2004). By 2002, quagga mussels were established in eight of nine reservoirs along the Volga River cascade (Orlova and Shcherbina, 2002). This recent, rapid spread is cause for concern as few taxa have as great a capability to transform ecosystem functioning and alter native species diversity (Karatayev et al, 1997; Ricciardi et al, 1998; Strayer et al, 1999). Even within the genus, it appears that strong interspecific interactions are possible. For example, the ability of quagga mussels to live in cold as well as warm water (Dermott and Munawar, 1993; Mills et al, 1996) has allowed this species to invade and displace zebra mussels in deeper water habitats in the lower Great Lakes (Stoeckmann, 2003). Other invertebrate species also have experienced shifts in abundance concomitant with quagga mussel establishment. Consequently, continued range expansion could have profound conservation implications for lakes, reservoirs and estuaries in both Eurasia and North America (Mills et al, 1993, 1996, 1999; Orlova and Shcherbina, 2002) highlighting the importance for the need to monitor and understand its dispersal dynamics.
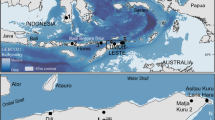
Location of Eurasian populations used for microsatellite analyses: (1) Caspian Sea; (2) Volga Delta; (3) Dniester Liman; (4) Bug Liman; (5) Dnieper Liman; (6) Kakhovka Reservoir; (7) Kubyshev Reservoir (Krasnovidovo); (8) Kubyshev Reservoir (Kama River); (9) Cheboksary Reservoir; (10) Uglich Reservoir; (11) Rybinsk Reservoir (Koprino); and (12) Rybinsk Reservoir (Mologa).
Despite a chronological record of establishment and the possibility of primary and secondary invasions, the mechanism(s) responsible for the rapid range expansion in Eurasia remain unknown. Human-mediated activities have facilitated dispersal, enabling the species to circumvent previously impassable barriers. Channelization and reservoir impoundment, over the past century, has created waterways navigable by commercial ships from the Baltic Sea to the Caspian Sea via the Volga River, with an additional connection to the Black-Azov Sea basin via the Don-Volga canal. In addition to the human-mediated dispersal, conversion of some stretches of the Volga River from riverine to lacustrine habitat appears to have facilitated the species' spread (Orlova and Shcherbina, 2002; Orlova et al, 2004). Once invasion pathways have been established between donor and recipient areas by major vectors (eg commercial ships), multiple seedings of individuals could occur. For example, it appears that establishment and spread of Chinese mitten crabs, Eriocheir sinensis, in central Europe resulted from repeated introductions (Hänfling et al, 2002). No previous genetic analyses have been undertaken to identify probable source populations or potential invasion pathways for quagga mussels in Eurasia.
In general, the application of molecular markers has proven useful to characterize dispersal patterns by NIS (eg Havel et al, 2000; Cristescu et al, 2001; Hänfling et al, 2002). However, factors that are responsible for NIS invasion success have been considerably less well studied (Lee, 2002). Previous surveys of allozymes and mitochondrial DNA have identified similar levels of genetic diversity between source and invading populations of Dreissena mussels (Hebert et al, 1989; Marsden et al, 1995; Wilson et al, 1999b; Lewis et al, 2000) and bivalves in general (Duda, 1994; Holland, 2001), suggesting a lack of founder or selection effects. In contrast, reduced genetic diversity has been reported for other invading organisms including waterfleas, Bythotrephes longimanus and Cercopagis pengoi (Berg and Garton, 1994; Cristescu et al, 2001; Berg et al, 2002; Therriault et al, 2002a, 2002b), the green alga Codium fragile spp. trementosoides (Provan et al, 2005) and the periwinkle Litorina saxatalis (Knight et al, 1987). Thus, contrasting observations exist with respect to invasion success and genetic diversity within invading populations.
Species may spread locally through population growth and regional diffusion, a process termed reaction-diffusion (Hengeveld, 1989). Alternatively, they may exhibit patterns of stratified-dispersal, involving both local diffusion and long-distance movement of propagules (Hengeveld, 1989; MacIsaac et al, 2002). Dreissena mussels have a biphasic life cycle involving veliger larvae and sedentary juveniles and adults, both of which contribute to dispersal. It appears that both reaction-diffusion and stratified diffusion have contributed to the spread of these dreissenid mussels (Wilson et al, 1999b; Johnson et al, 2001).
Analysis of time-series distribution data and genetic surveys may provide investigators with insights into dispersal mechanisms. For example, AFLP fingerprinting indicated that D. polymorpha, which invaded Ireland around 1994, entered as adults fouled on the hull of a pleasure boat imported from Britain (Pollux et al, 2003). In the Great Lakes basin, quagga mussels appear to have spread through Lakes Erie and Ontario via local diffusion (Mills et al, 1993). However, analyses of microsatellite markers for eight populations in these lakes indicated long-distance gene flow and a possible role of boaters in dispersal of the species (Wilson et al, 1999b). No population genetic studies have been conducted to characterize invasive quagga mussel populations in the Volga River system. The primary objective of this study was to combine invasion history information with population genetic data to identify the level of genetic diversity among invading and endemic, or other probable source, populations of quagga mussels in the Ponto-Caspian region and to identify potential genetic bottlenecks or founder effects. We used six highly polymorphic microsatellite markers to indirectly measure dispersal and population relatedness through the Volga River system, endemic populations in the Black Sea basin and a trans-oceanic introduction to North America. If the genetic data conform to the expectations of an isolation-by-distance model, one can infer that range expansion is occurring primarily by diffusion (eg Wright, 1943; Slatkin, 1993). Otherwise, stratified diffusion or jump-dispersal might be contributing (Wilson et al, 1999b).
Recently, introduced populations are expected to show reduced genetic diversity due to founder effects (eg Knight et al, 1987) but this is not always the case, especially for mussels (eg Hebert et al, 1989; Duda, 1994; Holland, 2001). Because we have sampled source and invaded populations, it should be possible to identify founder effects if they exist. Otherwise, populations may have been founded by large inocula or repeated introductions. Finally, we will comment on the claim that invasion success requires high genetic diversity and why this could be important for population persistence or the potential for further spread.
Materials and methods
Mussel taxonomy
Dreissenid molluscs have been reclassified many times and at many levels (ie genus, subgenus, species, subspecies and variety), based mainly on morphological, anatomical and geographical characteristics (see reviews by Babak, 1983; Rosenberg and Ludyanskiy, 1994; Starobogatov, 1994). The recent application of molecular techniques has helped to clarify the phylogenetic relationships among species (Spidle et al, 1994; Marsden et al, 1996; Claxton et al, 1998; Therriault et al, 2004). In the most recent analysis, Therriault et al (2004) proposed that D. bugensis was not sufficiently genetically distinct from D. rostriformis, a species native to the central and southern Caspian Sea, to be considered a separate species. Rather, they suggest that two races of D. rostriformis exist, one that lives in freshwater habitats (bugensis) and one that occurs in brackish waters (rostriformis=distincta). Consequently, throughout the rest of this paper, the quagga mussel will be referred to as D. rostriformis bugensis.
Study area and sample collection
Dreissena samples were collected from four locations in the Black Sea basin (two endemic and two invaded), seven recently invaded locations along the Volga River cascade, one native location in the central Caspian Sea and one introduced location in North America, Lake Erie (Table 1; Figure 1).
DNA isolation and PCR amplification
Total DNA was extracted from mantle muscle tissues of specimens preserved in 95% ethanol using either a standard phenol–chloroform method (Sambrook et al, 1989) or a DNA purification kit (Wizard, Promega). Extracted genomic DNA was used as a template for DNA amplification using the polymerase chain reaction (PCR). We amplified six microsatellite loci (Dbug 1–6 in Wilson et al, 1999a, 1999b) from at least 32 individuals from each of the 13 populations. PCR mixtures included double-distilled water, 10 × manufacturer-supplied PCR buffer, 25 mM MgCl2, 0.2 mM of each dNTP and 0.5 U Taq DNA polymerase (Gibco BRL). Reactions were run on a PTC-100 Programmable Thermal Controller (MJ Research Inc.) using an initial denaturation step at 95°C (5 min) followed by 40 cycles consisting of denaturation at 95°C (60 s), annealing at 49°C (60 s) and an extension step at 72°C (90 s). A final 7 min at 72°C was added after cycling to ensure complete extension. The annealing temperature used here is lower than the ones used by Wilson et al (1999a) in their development of these microsatellite primers but yielded the most consistent amplification with our reagents and sequencer.
Microsatellite DNA fragments were sized using manufacturer-supplied size standards on a CEQ8000 automated sequencer (Beckman Coulter). We assumed that alleles encountered in less than three individuals (0.7%) were amplification artifacts, possibly due to the lower annealing temperature, and these were not included in subsequent analyses.
Statistical analyses
We tested all populations by locus combinations for departure from Hardy-Weinberg expectations using Guo and Thompson's (1992) estimate in Arlequin version 2.0 software (Schneider et al, 2000): a 10 000-step, 1000-iteration Markov chain method provided an unbiased estimate of significance. A sequential Bonferroni correction was applied to account for the multiple simultaneous comparisons generated by these analyses (Rice, 1989). An exact test for differences in allele frequencies among populations was employed (20 000 permutations; Raymond and Rousset, 1995) using Tools for Population Genetic Analyses (TFPGA v 1.3) software by Mark Miller (Dept of Fisheries and Wildlife, Utah State University). Significance levels were subsequently adjusted using the sequential Bonferroni correction method (Rice, 1989).
Phylogenetic relationships among populations were inferred using the neighbor-joining (NJ) tree-building method in Populations 1.2.26 software by O Langella (Centre National de la Recherche Scientifique, Laboratoire Populations, Génétique et Evolution), using Nei's (1972) genetic distance DS. The tree was viewed using TreeView 1.6.6 software by Roderic Page (Institute of Biomedical and Life Sciences, University of Glasgow).
Pairwise genetic distances between populations were measured by calculating Nei's genetic distance DS (Nei, 1972) using TFPGA and FST using Arlequin. As populations subdivide (disperse or expand), models of isolation-by-distance provide one means to understand population expansion based on genetic distances. Thus, we plotted both Nei's genetic distance (DS) and FST vs geographical distances between populations (via waterways) to examine the relationship between geographical distance and genetic distance among populations. A Mantel test (10 000 permutations) was employed to statistically evaluate the correlation between genetic and geographic distance. These analyses were conducted using only Eurasian populations, to minimize the likelihood of spurious results associated with a single population from North America. As the Caspian Sea population was markedly genetically divergent from the other populations used in this study, it was also excluded from this analysis.
Results
We observed a total of 179 alleles from the six loci for the 421 individuals from 13 populations. We identified 36 alleles for Dbug1 (120–296), 34 alleles for Dbug2 (176–308), 33 alleles for Dbug3 (261–354), 29 alleles for Dbug4 (183–360), 36 alleles for Dbug5 (195–384) and 11 alleles for Dbug6 (165–273). Following Bonferroni correction, only the Lake Erie and Volga Delta pairwise comparison was not statistically significant. Further, exact tests showed that the Caspian Sea population was significantly different from all others (P<0.0001). Average heterozygosity was similar among endemic and invaded populations, suggesting that genetic diversity was not lower among invaded populations (Appendix 1 in supplementary information).
Globally, all six loci exhibited significant heterozygote deficiencies (P<0.0001). Intrapopulation comparisons revealed that heterozygote deficiencies were not significant for three populations at Dbug1, nine populations at Dbug3, one population at Dbug5 and two populations at Dbug6 following Bonferroni correction (Appendix 1 in supplementary information).
The Caspian Sea population was genetically distinct (Figure 2) particularly in the low number of alleles (5) identified for the Dbug2 locus (Appendix 1 in supplementary information). There was nonamplification in 25 of 32 individuals at this locus, which is consistent with a high frequency of null alleles in the population.
Unrooted NJ tree showing the relationship among 13 D. rostriformis populations using Nei's (1972) genetic distance. Bootstrap support based on 1000 iterations is shown for nodes with more than 70% support. Numbers in parentheses correspond to Map ID numbers in Table 1.
Allelic variation was too high to allow resolution between source and invaded populations. However, the relatively higher bootstrap support for the Dniester Liman – Cheboksary Reservoir node (77%) suggests a potential source – destination relationship (Figure 2). Based on invasion timing (Table 1), it is difficult to infer which population represents the source and which represents the destination. Further, the extremely low FST value between the Dniester Liman and Cheboksary Reservoir populations (0.0002) could indicate a high level of continuing gene flow (Appendix 2 in supplementary information). The Lake Erie population clustered with the Volga Delta population and to a lesser extent with the Dnieper Liman population, but bootstrap support was weaker (Figure 2). Since Lake Erie was invaded prior to the Volga Delta (Table 1), it appears that either the Lake Erie population was the source of the Volga Delta population or, alternatively, that the Dnieper Liman was the source for both invaded populations.
The analysis of pairwise FST indicated that neither endemic population (Bug Liman or Dnieper Liman) nor the invaded population in Rybinsk Reservoir at Koprino was the source for the Dniester Liman or Cheboksary Reservoir populations (Appendix 2 in supplementary information).
Correlations between geographical distance and DS and FST were not significant (Mantel test, P=0.1943 and 0.6644, respectively), even when only invaded populations were included in the analysis (P=0.1893 and 0.7013, respectively). Similarly, no relationships were detected when comparisons were made with only the two endemic populations (P=0.8249 and 0.6129, respectively).
Discussion
Colonization events may be accompanied by founder effects, in which the genetic diversity of a colonizing population is reduced relative to the source (eg Wright, 1943; Slatkin, 1993; Hänfling et al, 2002; see Holland, 2001 for contrary results). Alternatively, or in addition to this, genetic diversity could be reduced by postcolonization selection by the novel environment (eg Lee, 2002). In this study, we observed no evidence of reduced genetic diversity among introduced populations in either the Volga River or Lake Erie, a finding consistent with Wilson et al's (1999b) study on quagga mussels in the Great Lakes. Microsatellite diversity at the six polymorphic loci examined was similar across source and destination sites, indicating that inoculum sizes were large, there was swift population growth or that multiple introductions served to enhance existing genetic diversity in the invaded mussel populations. Although multiple introductions cannot be excluded for the Lake Erie population, the likelihood of this is lower than for invading populations in the Volga River system. An alternative explanation is that the Lake Erie introduction resulted from a very large propagule number that did not restrict genetic diversity, a hypothesis also applicable to invading populations in the Volga River system.
Population genetic theory predicts that high genetic diversity predisposes invasive populations to success at establishing, persisting, and dispersing further into novel habitats (Hartl and Clark, 1989). The quagga mussel populations surveyed in this study were characterized by high genetic diversity within populations with negligible population subdivision. High genetic diversity has been reported previously for Dreissena species (eg Marsden et al, 1995; Wilson et al, 1999b; Lewis et al, 2000) as well as other bivalves (eg Duda, 1994; Holland, 2001) and may be an important attribute contributing to their continued invasion success. Highly polymorphic populations are expected to retain maximum genetic diversity, providing genetic variation, which ensures that at least some individuals will be suited to survive and/or disperse to novel habitats. In contrast, a bottleneck would restrict genetic diversity and potentially lower the probability of successful establishment of an invading population. However, it should be noted that some highly successful invaders exhibit low genetic diversity. For example, successful invasive populations of the green alga C. fragile spp. trementosoides in the Mediterranean Sea, northern Europe, eastern North America, and in the South Pacific all possess markedly lower haplotype diversity relative to native populations in Japan (Provan et al, 2005). Similarly, introduced populations of Bythotrephes and Cercopagis in the Great Lakes have lower genetic diversity compared to native populations in Europe (Berg and Garton, 1994; Cristescu et al, 2001; Berg et al, 2002; Therriault et al, 2002a, 2002b). Further, environmental differences between the introduced and source habitats could result in selection and further restriction of genetic diversity (Lee, 2002). In our study, it appears that neither founder effects nor varying selection pressures affected genetic diversity of introduced quagga mussel populations. Although there is no consensus on whether successful invasions are characterized by reduced genetic diversity due to founder effects and selection or elevated genetic diversity reflecting multiple colonization events or large propagule size, our study is one of only a few that show no evidence for a founder effect or selection. Thus, high genetic diversity within the source population may be important for invasion success.
Historical accounts suggest two endemic, freshwater populations of quagga mussel from the Black Sea basin (Bug and Dnieper Limans; Table 1). As neither endemic population appears to have been the source for the Dniester Liman population (Figure 2), this population may have been established by a ‘reverse invasion’ from Cheboksary Reservoir. Further, since the Cheboksary Reservoir population seems not to have been established by an invasion from either endemic population, it may have resulted from a secondary invasion from a population established elsewhere, possibly within the Caspian Sea basin. The Volga Delta population and the Lake Erie population were not significantly different based on the pairwise exact tests and they clustered at the same node in the NJ tree (Figure 2). Thus, based on invasion timing, the Lake Erie population could represent the source of the Volga Delta population. Alternatively, a third population could represent the source of both populations. In fact, based on the position of the Dnieper Liman population in the NJ tree, we suggest that this population was responsible for both the Lake Erie and Volga Delta populations. This interpretation is consistent with a previous study based on allozyme variation that concluded that the quagga mussels in Lakes Erie and Ontario came from the lower Dnieper River (Spidle et al, 1994). Also, this finding is consistent with commercial shipping acting as the primary dispersal vector along known invasion corridors both within Eurasia and to North America (Ricciardi and MacIsaac, 2000; Orlova et al, 2004). Further, the current rapid spread of quagga mussels in Eurasia and North America began nearly simultaneously in the early 1990s (Table 1; Orlova et al, 2004), suggesting that a previously limiting barrier to dispersal was removed, although such a barrier has not been identified.
Many of the sampled populations showed significant departures from Hardy–Weinberg equilibrium at multiple loci, which is consistent with other studies of newly colonized bivalve populations (eg Wilson et al, 1999b) and may result from periodic re-introduction or strong selection effects. Alternatively, the departures from Hardy–Weinberg equilibrium could be due to null alleles (eg Pemberton et al, 1995) or a Wahlund effect (eg Hartl and Clark, 1989); however, neither explanation is expected to apply across all populations and loci (Castric et al, 2002). The Caspian Sea population was the only one for which the null allele hypothesis is supported by the occurrence of putative null homozygotes (ie nonamplification) at a single locus (Dbug2).
Our data suggest that sampled populations are part of a larger metapopulation that is able to maintain high levels of gene flow, possibly due to multiple colonization events between source and invaded populations. In common with numerous other studies of bivalves, we failed to identify a genetic pattern characteristic of isolation-by-distance. For example, Wilson et al (1999b) noted that quagga mussel populations in Lakes Erie and Ontario lacked an isolation-by-distance structure, possibly owing to long-distance dispersal of propagules by boats. Given the importance of the Volga River cascade as a commercial waterway for much of Eurasia, we suspect that vessel traffic has and will continue to play an integral role in dispersing quagga mussels throughout this system.
The mussel population from the Caspian Sea was the most divergent of the populations studied (Figure 2) and is the only population where there is convincing evidence of a high frequency of a null allele. Clearly, the Caspian Sea population did not found the populations in the Volga River system (or Black Sea basin) despite their closer geographical proximity. This is the only population known to inhabit brackish water and its divergence may reflect the spatial and temporal differentiation between the Black and Caspian Seas and different selection regimes encountered in these environments. The Black and Caspian basins last had contact approximately 7000–10000 years ago prior to the intrusion of Mediterranean waters into the Black Sea basin that displaced many brackish water species (Reid and Orlova, 2002). Therriault et al (2004) noted consistent but subtle genetic differences between quagga mussel populations from these basins, and argued that they represented two races, one that evolved in brackish water, D. rostriformis rostriformis (=distincta), and another that evolved in freshwater, D. rostriformis bugensis. Thus, populations in the Caspian Sea are presumed to have a greater salinity tolerance than those from freshwater estuaries of the Black Sea (Therriault et al, 2004). The recent arrival of the freshwater race to the Caspian Sea (Volga Delta population) could have significant conservation implications to the existing brackish water population in the Caspian Sea proper if contact between these to populations (races) occurs. In addition to the obvious genetic implications, there are ecological considerations. For example, displacement among Dreissena species has been reported in the Great Lakes and the Volga River system (Stoeckmann, 2003; Orlova et al, 2004). Thus, the encroachment of the freshwater race into the brackish water Caspian Sea could pose a conservation threat to the native population in less brackish sections of the Caspian Sea.
References
Antonov PI (1993). About invasion of bivalve Dreissena bugensis (Andr.) into the Volga River Reservoirs. Ecological problems of large rivers basins. Book of Abstracts of International Conference, Togliatti, Russia, 6–10 September, 1993. Publishers of IEVB RAS, Togliatti, Russia. pp 52–53 (in Russian).
Babak EV (1983). The Pliocene and Quaternary Dreissenidae of the Evsinsk Basin. Akad Nauk SSSR Tr Paleontol Inst 204: 1–104 (in Russian).
Berg DJ, Garton DW (1994). Genetic differentiation in North American and European populations of the cladoceran Bythotrephes. Limnol Oceanogr 39: 1503–1516.
Berg DJ, Garton DW, MacIsaac HJ, Panov VE, Telesh IV (2002). Changes in genetic structure of North American Bythotrephes populations following invasion from Lake Ladoga, Russia. Freshwater Biol 47: 275–282.
Castric V, Bernatchez L, Belkhir K, Bonhomme F (2002). Heterozygote deficiencies in small lacustrine populations of brook charr Salvelinus fontinalis Mitchell (Pisces, Salmonidae): a test of alternative hypotheses. Heredity 89: 27–35.
Claxton WT, Wilson AB, Mackie GL, Boulding EG (1998). A genetic and morphological comparison of shallow- and deep-water populations of the introduced dreissenid bivalve Dreissena bugensis. Can J Zool 76: 1269–1276.
Cristescu MEA, Hebert PDN, Witt DS, MacIsaac HJ, Grigorovich IA (2001). An invasion history for Cercopagis pengoi based on mitochondrial gene sequences. Limnol Oceanogr 46: 224–229.
Dermott R, Munawar M (1993). Invasion of Lake Erie offshore sediments by Dreissena, and its ecological implications. Can J Fish Aquat Sci 50: 2298–2304.
Duda Jr TF (1994). Genetic population structure of the recently introduced Asian clam, Potamocorbula amurensis, in San Francisco Bay. Mar Biol 119: 235–241.
Guo SW, Thompson EA (1992). A Monte-Carlo method for combined segregation and linkage analysis. American Journal of Human Genetics 51: 1111–1126.
Hänfling B, Carvalho GR, Brandl R (2002). mt-DNA sequences and possible invasion pathways of the Chinese mitten crab. Mar Ecol Prog Ser 238: 307–310.
Hartl DL, Clark AG (1989). Principles of Population Genetics. Sinauer Associates Inc.: Sunderland, MA.
Havel JE, Colbourne JK, Hebert PDN (2000). Reconstructing the history of intercontinental dispersal in Daphnia lumholtzi by use of genetic markers. Limnol Oceanogr 45: 1414–1419.
Hebert PDN, Muncaster BW, Mackie GL (1989). Ecological and genetic studies on Dreissena polymorpha (Pallas): a new mollusc in the Great Lakes. Can J Fish Aquat Sci 46: 1587–1591.
Hengeveld R (1989). Dynamics of Biological Invasions. Chapman & Hall: NY.
Holland BS (2001). Invasion without a bottleneck: microsatellite variation in natural and invasive populations of the brown mussel Perna perna (L). Mar Biotechnol 3: 407–415.
Johnson LE, Carlton JT, Ricciardi A (2001). Overland dispersal of aquatic invasive species: a risk assessment of transient recreational boating. Ecol Appl 11: 1789–1799.
Karatayev AY, Burlakova LE, Padilla DK (1997). The effects of Dreissena polymorpha (Pallas) invasion on aquatic communities in Eastern Europe. J Shellfish Res 16: 187–203.
Kharchenko TA (1995). Dreissena: range, ecology, biofouling. Gidrobiologicheskii Zh 31: 3–20 (in Russian).
Kinzelbach R (1992). The main features of the phylogeny and dispersal of the zebra mussel Dreissena polymorpha. Limnol Aktuell 4: 5–17.
Knight AJ, Hughes RN, Ward RD (1987). A striking example of the founder effect in the mollusc Litorina saxatalis. Biol J Linn Soc 32: 417–426.
Lee CE (2002). Evolutionary genetics of invasive species. Trends Ecol Evol 17: 386–391.
Lewis KM, Feder JL, Lamberti GA (2000). Population genetics of the zebra mussel, Dreissena polymorpha (Pallas): local allozyme differentiation within Midwestern lakes and streams. Can J Fish Aquat Sci 57: 637–643.
MacIsaac HJ, Robbins TC, Lewis MA (2002). Modeling ships' ballast water as invasion threats to the Great Lakes. Can J Fish Aquat Sci 59: 1245–1256.
Marsden JE, Spidle AP, May B (1995). Genetic similarity among zebra mussel populations within North America and Europe. Can J Fish Aquat Sci 52: 836–847.
Marsden JE, Spidle AP, May B (1996). Review of genetic studies of Dreissena spp. Am Zool 36: 259–270.
May B, Marsden JE (1992). Genetic identification and implications of another invasive species of Dreissenid mussel in the Great Lakes. Can J Fish Aquat Sci 49: 1501–1506.
Minchin D, Lucy F, Sullivan M (2002). Zebra mussel: impact and spread. In: Leppäkoski E, Gollasch S, Olenin S (eds) Aquatic Invasive Species of Europe. Distribution, Impacts and Management. Kluwer Publishers: Netherlands, pp 135–146.
Mills EL, Chrisman JR, Baldwin B, Owens RW, O'Gorman R, Howell T et al (1999). Changes in the dreissenid community in the Lower Great Lakes with emphasis on southern Lake Ontario. J Great Lakes Res 25: 187–197.
Mills EL, Dermott RM, Roseman EF, Dustin D, Mellina E, Conn DB et al (1993). Colonization, ecology, and population structure of the ‘quagga’ mussel (Bivalvia: Dreissenidae) in the lower Great Lakes. Can J Fish Aquat Sci 50: 2305–2314.
Mills EL, Rosenberg G, Spidle AP, Ludyanskiy M, Pligin Y, May B (1996). A review of the biology and ecology of the quagga mussel (Dreissena bugensis), a second species of freshwater dreissenid introduced to North America. Am Zool 36: 271–286.
Nei M (1972). Genetic distance between populations. Am Nat 106: 283–291.
Orlova MI, Muirhead J, Antonov PI, Shcherbina GKh, Starobogatov YI, Biochino GI et al (2004). Range expansion of quagga mussels Dreissena rostriformis bugensis in the Volga River and Caspian Sea basin. Aquat Ecol 38: 561–573.
Orlova MI, Shcherbina GH (2002). About range expansion of Dreissena bugensis Andr. Dreissenidae, Bivalvia) in Upper Volga Reservoirs. Zool J 81: 515–520 (in Russian).
Pemberton JM, Slate J, Bancroft DR, Barrett JA (1995). Nonamplifying alleles at microsatellite loci: a caution for parentage and population studies. Mol Ecol 4: 249–252.
Pollux B, Minchin D, Van der Velde G, Van Alen T, Moon-Van Der Staay SY, Hackstein J (2003). Zebra mussels (Dreissena polymorpha) in Ireland, AFLP-fingerprinting and boat traffic both indicate an origin from Britain. Freshwater Biol 48: 1127–1139.
Provan J, Murphy S, Maggs CA (2005). Tracking the invasive history of the green alga Codium fragile spp trementosoides. Mol Ecol 14: 189–194.
Raymond M, Rousset F (1995). An exact test for population differentiation. Evolution 49: 1280–1283.
Reid DF, Orlova MI (2002). Geological and evolutionary underpinnings for the success of Ponto-Caspian species invasions in the Baltic Sea and North American Great Lakes. Can J Fish Aquat Sci 59: 1144–1158.
Ricciardi A, MacIsaac HJ (2000). Recent mass invasion of the North American Great Lakes by Ponto-Caspian species. Trends Ecol Evol 15: 62–65.
Ricciardi A, Never RJ, Rasmussen JB (1998). Impending extinctions of North American freshwater mussels (Unionida) following zebra mussel (Dreissena polymorpha) invasion. J Anim Ecol 67: 613–619.
Rice WR (1989). Analyzing tables of statistical tests. Evolution 43: 223–225.
Rosenberg G, Ludyanskiy ML (1994). A nomenclatural review of Dreissena (Bivalvia: Dreissenidae), with identification of the quagga mussel as Dreissena bugensis. Can J Fish Aquat Sci 51: 1474–1484.
Sambrook J, Fritsch EF, Maniatis T (1989). Molecular Cloning? A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY.
Schneider S, Roessli D, Excoffier L (2000). Arlequin ver. 2.000: A Software for Population Genetics Data Analysis. Genetics and Biometry Laboratory, University of Geneva: Switzerland.
Slatkin M (1993). Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47: 264–279.
Spidle AP, Marsden JE, May B (1994). Identification of the Great Lakes quagga mussel as Dreissena bugensis from the Dnieper River, Ukraine, on the basis of allozyme variation. Can J Fish Aquat Sci 51: 1485–1489.
Starobogatov YI (1994). Freshwater Zebra Mussel, Dreissena polymorpha (Pall.) (Bivalvia, Dreissenidae): Systematics, Ecology, Practical Meaning, Species of the Fauna of Russia and Adjacent Countries. Nuaka: Moscow. 240pp (in Russian).
Stoeckmann A (2003). Physiological energetics of Lake Erie dreissenid mussels: a basis for the displacement of Dreissena polymorpha by Dreissena bugensis. Can J Fish Aquat Sci 60: 126–134.
Strayer DL, Caraco NF, Cole JJ, Findlay S, Pace ML (1999). Transformation of freshwater ecosystems by bivalves. BioScience 49: 19–27.
Therriault TW, Docker MF, Orlova MI, Heath DD, MacIsaac HJ (2004). Molecular resolution of Dreissenidae (Mollusca: Bivalvia) including the first report of Mytilopsis leucophaeta in the Black Sea basin. Mol Phylogenet Evol 30: 479–489.
Therriault TW, Grigorovich IA, Cristescu ME, Ketelaars HAM, Viljanen M, Heath DD et al (2002a). Taxonomic resolution of the genus Bythotrephes Leydig using molecular markers and re-evaluation of its global distribution. Diversity and Distributions 8: 67–84.
Therriault TW, Grigorovich IA, Kane DD, Haas EM, Culver DA, MacIsaac HJ (2002b). Range expansion of the exotic zooplankter Cercopagis pengoi in the Great Lakes. J Great Lakes Res 28: 698–701.
Wilson AB, Boulding EG, Naish K-A (1999a). Characterization of tri- and tetranucleotide microsatellite loci in the invasive mollusc Dreissena bugensis. Mol Ecol 8: 692–693.
Wilson AB, Naish K-A, Boulding EG (1999b). Multiple dispersal strategies of the invasive quagga mussel (Dreissena bugensis) as revealed by microsatellite analyses. Can J Fish Aquat Sci 56: 2248–2261.
Wright S (1943). Isolation by distance. Genetics 28: 114–138.
Acknowledgements
We are grateful for financial support in the form of an NSERC postdoctoral fellowship (TWT), GLIER postdoctoral fellowship (MFD), NSERC International Opportunities Fund (HJM), NSERC discovery grants (DDH and HJM), and Russian Foundation for Basic Research Project # 1634.2003.4 and Russian Federal Biodiversity Program (MIO).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Heredity website (http://www.nature.com/hdy)
Supplementary information
Rights and permissions
About this article
Cite this article
Therriault, T., Orlova, M., Docker, M. et al. Invasion genetics of a freshwater mussel (Dreissena rostriformis bugensis) in eastern Europe: high gene flow and multiple introductions. Heredity 95, 16–23 (2005). https://doi.org/10.1038/sj.hdy.6800691
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800691
Keywords
This article is cited by
-
Dreissenids’ breaking loose: differential attachment as a possible driver of the dominance shift between two invasive mussel species
Biological Invasions (2021)
-
Differential tolerance to nickel between Dreissena polymorpha and Dreissena rostriformis bugensis populations
Scientific Reports (2018)
-
Where can introduced populations learn their tricks? Searching for the geographical source of a species introduction to the Galápagos archipelago
Conservation Genetics (2017)
-
Two African origins of naturalized brown mussel (Perna perna) in Brazil: past and present bioinvasions
Hydrobiologia (2017)
-
Validated methodology for quantifying infestation levels of dreissenid mussels in environmental DNA (eDNA) samples
Scientific Reports (2016)