Abstract
South American Creole cattle are direct descendants of the animals brought to the New World by the Spanish and Portuguese during the 16th century. A portion of the mitochondrial D-loop was sequenced in 36 animals from five Creole cattle populations in Argentina and four in Bolivia. Individuals belonging to the potentially ancestral Spanish breed Retinta were also analysed. Sequence comparisons revealed three main groups: two with the characteristics of European breeds and a third showing the transitions representative of the African taurine breeds. The African sequences were found in two populations from Argentina and three populations from Bolivia, whose only connections go back to colonial times. The most probable explanation for the finding is that animals could have been moved from Africa to Spain during the long-lasting Arabian occupation that started in the seventh century, and from the Iberian Peninsula to America eight centuries later. However, since African haplotypes were not found in the Spanish sample, the possibility of cattle transported directly from Africa cannot be disregarded.
Similar content being viewed by others
Introduction
Cattle were introduced to the American continent by the Spanish Conquerors, who transported the first animals to the Caribbean Islands after the first trip of Columbus in 1492. The founding population consisted of approximately 300 Iberian cattle (Primo, 1992), which spread from Central America all over the continent. Argentinean Creole Cattle (ACc) and Bolivian Creole Cattle (BCc) are humpless bovines with long horns descendant from these cattle introduced between 1555 and 1587. After their arrival, they rapidly became adapted to a wide range of environments, from rainforest to Patagonian steppe in Argentina, and from tropical overflowed plain to Andean highland plain in Bolivia. During the 19th and 20th centuries, as a result of the introduction of highly selected European and Zebu commercial breeds, populations of Creole cattle suffered a severe reduction in number, and their distribution became restricted to marginal regions. Nowadays, ACc are distributed in semi-isolated populations of few individuals and low levels of gene flow (Giovambattista et al, 2001). They have been bred in semiwild conditions with low levels of artificial selection almost from their origin, suggesting that their genetic characteristics probably reflect very closely the original cattle brought to America from Spain.
Cattle in the Iberian Peninsula belong to two main groups (Alderson, 1981; Felius, 1985). The first group comprises the solid-coloured breeds of Central Europe, which is mainly distributed in the North West of Spain and Portugal. Cattle taken by the Portuguese conquerors to Brazil belonged to this group (Felius, 1985; Rodero et al, 1992). The second group includes the longhorned breeds of Western Europe, which corresponds to the Celtic breeds located mainly in Southwest Iberia (Alderson, 1981). It is assumed that cattle taken to America by the Spanish belonged to this second group, with most of the animals originating in Andalucia. Some historical records also describe cattle taken to America from the Canary Islands, a very common intermediate port used by Spanish ships going to the New World (Primo, 1992; Rodero et al, 1992). However, there is evidence that pre-Hispanic cattle did not exist in the archipelago, and the animals found there may have had their origin in Andalucia (Rodero et al, 1992).
Martin-Burriel et al (1999) recognised a third type of Iberian cattle, the so-called Black Orthoide, which they characterise as an example of ancient wild cattle present in the Iberian Peninsula before the other breeds arrived, and actually located in Central Spain. These cattle would be the result of local domestication of wild oxen (Sanchez-Belda, 1984).
Many recent studies have focused on the differentiation between European and African cattle. In this respect, mitochondrial DNA (mtDNA) has been a very valuable tool in the understanding of the origin and domestication of cattle. The pioneer studies of Loftus et al (1994a),(1994b) and Bradley et al (1996) showed the existence of two main groups of haplotypes, representatives of Bos indicus (zebu) and Bos taurus (taurine) lineages. They also identified independent domestication events for both strains, with zebu cattle domesticated in Asia and taurine animals from a separate group of aurochs in the Near East and Africa (Loftus et al, 1994a). More extensive studies of taurine breeds in Europe, Africa and the Near East showed that almost all sequences have their roots in one of four main ancestral haplotypes, one of them being the predominant haplotype in Africa and another in Europe and the Near East. Each geographic location sampled produced a star-like phylogeny centred on one of these two haplotypes, which differ only in transitions at positions 16050, 16113 and 16255 of the mitochondrial D-loop. The results also suggested a separate African domestication of taurine cattle. This mtDNA-based finding was also confirmed with microsatellite studies (MacHugh et al, 1997; Loftus et al, 1999; Hanotte et al, 2002), which revealed that the earlier cattle in Africa originated within the continent and suffered later introgression from Europe and the Near East.
This paper presents the first mtDNA characterisation of Argentinean and Bolivian Creole cattle, and reports the existence of African as well as European haplotypes in cattle from South America.
Materials and methods
Study sites and sample collection
Blood samples were collected from 181 adult animals from nine populations throughout the range of distribution of ACc and BCc (Figure 1). When pedigree information was available, individuals that have not shared a common ancestor for at least three generations were selected. The populations sampled were chosen to represent a variety of environments inhabited by Creole cattle: in Bolivia, subtropical dry forest (Bolivian Chaqueño breed, Estación Experimental El Salvador), tropical overflow plain (Yacumeño breed, Departamento de El Beni), tropical plain (Saavedreño breed, Estación Experimental de Saavedra) and Andean highland plain (Chusco breed, Patacamaya). In Argentina the populations included habitats such as temperate plain (Balcarce, Tandileufú), dry forest (El Remate, Arroyo del Medio) and Andean cold forest (Calafate). Two Brahman cattle from Bolivia and five animals belonging to the Retinta breed from Spain were also included in the analysis. Retinta is the most numerous breed in Spain, and its distribution is centred in Western Andalucia and Extremadura.
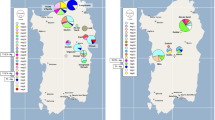
Map showing the populations of Argentinean and Bolivian cattle examined. The proportion of African haplotypes in each sample is illustrated by the filled sectors of the pie charts. YCc: Yacumeño breed, ChuCc: Chusco breed, SCc: Saavedreño breed, ChCc: Bolivian Chaqueño, ERCc: El Remate, AMCc: Arroyo del Medio, BCc: Balcarce, TCc: Tandileufú, CCc: Calafate.
DNA extraction, PCR amplification and sequencing of the D-loop
Total DNA was extracted from blood samples using the DNAZOL purification kit (Gibco Life Technologies, Rockville, MD, USA) following the manufacturer's instructions. Animals were preselected before sequencing by PCR-SSCP analysis to include a range of genetically distinct individuals. The PCR primers used were SSCPG3.F and SSCPG4.R (Eledath and Hines, 1996), which amplify a 489-base-pair fragment of the D-loop. The 50 μl reaction mix contained approximately 100 ng of total DNA, 0.5 μM of each primer, 0.1 mM of dNTPs and 2 U of Taq polymerase (Gibco Life Technologies, Rockville, MD, USA) in 20 mM Tris-HCl (pH 8.4), 50 mM KCl and 2 mM MgCl2, under mineral oil. The PCR consisted of a first denaturation step at 94°C for 2 min followed by 30 cycles of 45 s at 94°C, 1 min at 50°C and 1 min at 72°C, with an elongation step of 5 min at 72°C in the last cycle. For the SSCP analysis, 15 μl microliters of each PCR product was added to 40 μl of dye LIS (10% sucrose, 0.01% bromophenol blue and 0.01% xylene cyanol FF). The samples were then heated at 96°C for 10 min, cooled on ice for at least 5 min and loaded onto a 10% polyacrylamide gel (38:1 acrylamide:bisacrylamide). Electrophoresis was carried out at 4°C, 200 V in 0.5 × TBE buffer for 16 h. The gels were subsequently fixed in 5% ethanol, stained with 0.2% AgNO3 and revealed with 2% CaCO3.
PCR products were then purified using QIAquick columns (QIAGEN) and cloned into dT-tailed pGEM-T easy vector (Promega, Madison, WI, USA). DNA sequencing was performed in an Applied Biosystems 377 automated sequencer using T7 and M13 universal primers. At least two independent clones were sequenced from each individual and the unique substitutions were confirmed by sequencing clones from a second PCR product. In order to corroborate the existence of particular mutations, a second fragment was sequenced in selected samples with primers L15960 and H16334 (Cymbron et al, 1999).
Data analysis
Sequences of the D-loop were aligned using CLUSTAL-V multiple alignment software (Higgins et al, 1992). Sites representing a gap in any of the aligned sequences were excluded from the analysis, and distances between D-loop sequences were estimated using both the absolute number of nucleotide differences and the Kimura two-parameter distance (Kimura, 1980) calculated on the basis of an equal substitution rate per site. Analysis of molecular variance (AMOVA) and pairwise FST distances were calculated using the Arlequin analysis package (Schneider et al, 1997). A reduced median network was constructed using the methodology described in Bandelt et al (1995). Phylogenies were produced using maximum parsimony with the PAUP 4.0 software (Swofford, 1997) and the NEIGHBOR program incorporated in the PHYLIP package (Felsenstein, 1991). The statistical confidence of each node in the consensus trees was estimated by 1000 bootstrap resampling of the data. All sequences have been submitted to GenBank (accession numbers AF531381–AF531413)
Results
Variation in the mitochondrial D-loop
A total of 181 cattle were examined with SSCP, 97 belonging to the Argentinean Creole and 84 to the Bolivian Creole breeds. PCR products were of the same length and heteroplasmy was not detected in the sample examined. The SSCP analysis revealed eight variants, all of them shared between different populations. Individuals representing different SSCP patterns and different populations were chosen for sequencing. At least one representative of each population was sequenced for each one of the variants. When the resulting sequences were not identical for any particular SSCP pattern, more individuals with the same pattern were sequenced. All SSCP patterns resulted in at least two different haplotypes, with the extreme case of five different haplotypes showing an indistinguishable SSCP pattern, differing in up to 12 nucleotides. This result clearly indicates the lack of utility of the SSCP method to distinguish between cattle mtDNA haplotypes.
A total of 36 animals from five Argentinean and four Bolivian populations were sequenced with primers SSCPG3.F and SSCPG4.R (Eledath and Hines, 1996).
In the 419 base pairs obtained, there were a total of 35 polymorphic sites characterising 23 different haplotypes in the Creole cattle (Table 1). There were 30 transitions, four transversions and one/two insertions located in a poly (C) stretch, relative to the bovine reference sequence (V00054, Anderson et al, 1982). The pattern of variation was comparable with those described previously for cattle. No indication of a bottleneck effect due to the founding of the populations in America was evident from haplotype diversity. The mean nucleotide diversity was 0.058 (SD 0.03) and the mean number of pairwise differences was 2.61 (SD 1.42). This is comparable with that observed in the European mainland (1.92), Britain (2.68) and Africa (2.09), and lower than the values for the Middle East (3.97) and Anatolia (3.49) (Troy et al, 2001). Apparently, the very low levels of artificial selection in the South American Creole breeds must have compensated for the bottleneck effect that would otherwise have reduced variability during the foundation of the American populations.
One Argentinean Holstein, two Brahman (B. indicus) and five Retinta animals were also sequenced, and resulted in four more haplotypes. The Retinta sample presented two different haplotypes, one unique (R2) and the other (R1) found in four animals. Argentinean Holstein showed a unique haplotype (ARHOL). The Brahman cattle sampled here showed taurine mtDNA, even when is described as a typical zebu breed very common in North America. One of the haplotypes was identical to one Creole haplotype (CC1), while the other was unique (BRA). This finding was not completely unexpected, since taurine mtDNA was also identified in 23 Brahman animals examined by RFLP (Meirelles et al, 1999). Zebu cattle were brought from India to North America at the beginning of the 19th century. There are records of less than 300 imported Brahmans, most of which were bulls, so it could be assumed that other breeds, possibly taurine, supplied the foundation animals for the breed in America (http://www.ansi.okstate.edu/breeds/cattle).
Within the 23 Creole haplotypes, CC1 was represented eight times (including the Brahman animal), CC8 seven times, five haplotypes (CC2, CC3, CC9, CC10 and CC18) were represented twice and the rest were detected only once.
Within the fragment analysed, a T/C transition at position 16255 separates African and European haplotypes of B. taurus, with C characterising cattle of African origin. This substitution was found in seven animals (CC18 toCC23) from two populations in Argentina (Calafate, El Remate, Figure 1) and three from Bolivia (Patacamaya, El Beni, Saavedra, Figure 1). To confirm their African origin, a second fragment was amplified from positions 15960 to 16334, in order to assess nucleotide variation at the two other diagnostic positions (16050 and 16113). The seven animals showed the characteristic African substitutions at those variable sites. Also, five typically European haplotypes were sequenced for the second fragment, with none showing the African transitions.
Although most of the rest of the substitutions were unique, position 169 showed an A/G transition, which separates the European group of sequences in two subgroups. This substitution has been previously described in many European and Asian breeds, and it could reflect the retention of ancestral polymorphism in the Near East founding population or the existence of at least two ancestral populations.
Previous studies based on polymorphism at loci related to milk production and immune response and on microsatellites have detected genetic structure among the Argentinean and Bolivian populations of cattle (Lirón et al, 2000; Giovambattista et al, 2001). An AMOVA based on the mitochondrial sequences was conducted in order to analyse the distribution of this variability. The results indicated that there is no genetic structure among the different populations examined (FST 0.021, nonsignificant).
Phylogenetic analysis
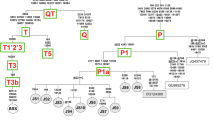
Figure 2 shows a network phylogeny constructed by hand as detailed in Bandelt et al (1995). Representatives of the published European and African haplotypes were added to the data set. All haplotypes fall within three main clusters: two of them representing sequences of European origin and the third group sequences of African origin. The star-like pattern observed is typical of all cattle phylogenies, and denotes a history of population expansion linked to domestication (Troy et al, 2001). The most frequent haplotypes are found in the centre of each cluster, which indicates that they are probably ancestral. This is confirmed by the fact that haplotype CC1 is identical to the reference sequence (V00054, Anderson et al, 1982) and to haplotype T3 in Troy et al (2001), while haplotype CC8 is also found in breeds such as Brown Swiss, Cannadiene and Japanese Black. The African consensus and more frequent haplotype (T1 in Troy et al, 2001) was not found in the Creole Cattle. This could indicate the result of two bottlenecks: one during the foundation of the Iberian cattle from African animals and a second one during the foundation of the American populations of cattle.
A median network with the mtDNA haplotypes found in the cattle analysed. Circles represent haplotypes, with area proportional to their frequency. Shaded circles represent sequences of African origin. Black dots denote theoretical intermediate nodes introduced by the algorithm. Branches between haplotypes represent substitutions labelled according to the reference sequence, underlined numbers indicate recurrent mutations. Numbers 1–23 represent Creole haplotypes, the other haplotypes reported here are ARHOL (Argentinean Holstein), BRA (Brahman), and R (Retinta). Representatives of European and African groups included: LIM (Limousine, AF034446), RA (Red Angus, AF034445), ND (ŃDama, L27731), BU (Butana, L27714). Haplotype 1 is identical to the reference sequence (V00054).
Phylogenetic trees were also constructed using maximum parsimony and distance methods (not shown). The topologies obtained, based on only six shared substitutions, some of which are probably homoplasies, were identical to the network of Figure 2. However, the bootstrap values of all nodes were always lower than 50%.
Discussion
Cattle were introduced to America by the Spanish and Portuguese conquerors during the 16th century. Although initially introduced in small numbers, the animals became widely distributed and very well adapted to the new environments. Spanish conquerors took the cattle to Central America and the Spanish colonies in South America, while the Portuguese transported animals to Brazil.
There is some debate about the origins of Iberian cattle. Although they are supposed to share ancestry with many other breeds from Europe, which were derived from primitive domestication centres in the Near East, it has also been suggested that the wild oxen populations of Iberia could have been domesticated locally (Davidson, 1989; Martin-Burriel et al, 1999). However, no conclusive genetic or archaeological evidence to support this has yet been found. It has also been postulated that, given the intense trading between the Balearic Islands, the Iberian Peninsula and the North of Africa, Iberian cattle could also reflect the influence of African breeds (Martin-Burriel et al, 1999). This influence was detected using mtDNA sequences in Portuguese cattle (Cymbron et al, 1999). The authors in this case attribute the presence of African haplotypes to the Arabian invasion of the Iberian Peninsula during the seventh century.
This paper reports the presence of African haplotypes in South American Creole cattle. An early indication of the influence of African breeds in the origin of Creole cattle is given in Giovambattista et al (2001), which describes the existence of two particular alleles of BoLA-DRB3 in Argentinean Creole cattle that were never detected in European breeds, but were reported in African indigenous breeds.
Different hypotheses can be put forward to explain the existence of African haplotypes in South American cattle. First, it could be the result of recent introgression of cattle from Brazil. Although no studies of mtDNA of Brazilian cattle have been published, it is possible that because African haplotypes are present in Portuguese cattle, they should also be present in the cattle from Brazil. The highest frequency of African haplotypes in Portugal was found in the southernmost breeds (Cymbron et al, 1999), and that region was the starting point for the trips to the New World. However, there is no evidence of imported Brazilian cattle to Argentina, except for sporadic cattle stealing during the Argentina–Brazil–Uruguay war at the beginning of the 19th century. African haplotypes were found in 19.4% of the Argentinean and Bolivian cattle and in five different populations. In Argentina, they were found in Patagonian cattle (Calafate) and in one population from the Northwest region (El Remate). Both populations represent relics of the original cattle brought by the Spanish conquerors, which survived isolated for almost 500 years and without crossbreeding with any other European cattle. This is also the case of the Bolivian populations where the African haplotypes were found. The haplotypes are too widespread to be the product of recent introgression, and the records show that there was no contact between those populations since the first introduction of cattle to South America.
A second possibility is that animals of African origin were taken to America by the Spanish conquerors directly from Africa or from the intermediate ports used during the trips. During the 15th century, Spain had colonies in the North of Africa, and it is possible that, as well as slaves, animals were transported from that region. Another possibility is that, given that animals probably died during the 2-month trips, they were replaced by new animals from the intermediate ports. Spanish sailors stopped and restock in the Canary Islands, colonised by Spain (Parry, 1970). The Canaries formed a kind of ‘Atlantic Mediterranean’ with the other groups of islands in the region–Madeira, Azores and Cape Verde, under Portuguese power (Johnson, 1987). Their economies were linked together by the sea. None of the islands had indigenous cattle (Rodero et al, 1992), but it was common practice to introduce animals to proliferate in the new surroundings before settlers arrived (Johnson, 1987). These animals, including cattle, may have originated from the North African Spanish colonies or the West African Portuguese colonies, and could have been transported later on to the New World, and with them the African haplotypes found now in South America (Figure 3).
A third possibility is that cattle of African origin were already present in Spain at the time of the Conquest. It has been proposed that the founding population of Iberian cattle may have been partially of African origin. The expansion of livestock may have proceeded following a Mediterranean route instead of through the continental forests (Davidson, 1989; Cymbron et al, 1999). Alternatively, the Arabian Moors could have introduced the animals during the 700-year occupation of the peninsula. They certainly introduced new crops and other domestic animals such as horses and sheep. Their influence was more important in the south of Iberia, which is in agreement with the higher proportion of African haplotypes being found in Portuguese cattle (Cymbron et al, 1999). It is important to note that the southern ports, such as Lisbon, Seville and Cadiz monopolised the navigation to America, and that is generally assumed that the exported animals came from the same areas as the explorers (Rodero et al, 1992).
The present work failed to find African haplotypes in the Spanish sample examined, although only five animals representative of the Retinta breed could be sequenced, and they showed only two different haplotypes. Nevertheless, representatives of the Berrenda breed from Spain were included in the study by Troy et al (2001). They found 11 different haplotypes, all of them belonging to the European group, which is in agreement with our results and do not support the hypothesis of African influence in the cattle from Spain.
While our mitochondrial data do not allow us to discriminate between an Arabian-mediated transport of animals to Spain during the occupation of Iberia and a transport of animals by the Spanish directly from Africa to America, other genetic studies may provide some clues about the source of African haplotypes in South American cattle. Modern African cattle have a substantial introgression of B. indicus genes (Bradley et al, 1994; MacHugh et al, 1997; Hanotte et al, 2002), including a high frequency of acrocentric Y chromosome characteristic of zebu. This Y morphology has never been found in cattle from Spain (e.g. Arruga and Zaragoza, 1985; Moreno-Millán et al, 1991) or Argentina (De Luca et al, 1997; Giovambattista et al, 2000), although it has been found in taurine cattle of Bolivia, probably due to a recent introgression of zebu from Brazil. Analysis of a polymorphic Y microsatellite also showed no zebuine-specific allele in the Argentinean cattle, with Bolivian populations showing between 17 and 41% of zebu introgression (Giovambattista et al, 2000). B. indicus seems to have moved in great numbers to Africa with the Arab settlements along the East from the end of the seventh century (Epstein, 1971). If the animals transported to America during the 16th Century came directly from Africa, they would probably show some of the indicine introgression that occurred in that continent. This last reflection would indicate that, as well as producing important cultural changes, the Arab conquest of Spain would be the source of African haplotypes in Creole cattle from South America.
References
Alderson L (1981). The categorisation of types and breeds of cattle in Europe. Arch Zootec 41: 325–334.
Anderson S, de Bruijn MH, Coulson AR, Eperon IC, Sanger F, Young IG (1982). Complete sequence of bovine mitochondrial DNA. J Mol Biol 156: 683–717.
Arruga MV, Zaragoza I (1985). Chromosome studies in Spanish fighting bulls. Arch Zootec 34: 49–65.
Bandelt HJ, Forster P, Sykes BC, Richards MB (1995). Mitochondrial portraits of human populations using median networks. Genetics 141: 743–753.
Bradley DG, MacHugh DE, Cunningham EP, Loftus RT (1996). Mitochondrial diversity and the origins of African and European cattle. Proc Natl Acad Sci USA 93: 5131–5135.
Cymbron T, Loftus RT, Malheiro MI, Bradley DG (1999). Mitochondrial sequence variation suggests an African influence in Portuguese cattle. Proc R Soc Lond B 266: 597–603.
Davidson I (1989). Escaped domestic animals and the introduction of agriculture in Spain. In: Clutton-Brock J (ed) The Walking Larder: Patterns of Domestication, Pastoralism and Predation, Unwin Hyman: London. pp 59–71.
De Luca JC, Golijow CD, Giovambattista G, Diessler M, Dulout FN (1997). Y-chromosome morphology and incidence of the 1/29 translocation in Argentine Creole bulls. Theriogenology 47: 761–764.
Eledath FM, Hines HC (1996). Detection of nucleotide variations in the D-loop region of bovine mitochondrial DNA using polymerase chain reaction-based methodologies. Anim Genet 27: 333–336.
Epstein H (1971). The Origin of Domestic Animals of Africa. Africana: New York.
Felius M (1985). Genus Bos: Cattle Breeds of the World. MSD AGVET, Merck and Co., Inc.: Rahwy, NJ.
Felsenstein J (1991). PHYLIP (phylogeny inference package), version 3.4. University of Washington: Seattle, WA.
Giovambattista G, Ripoli MV, De Luca JC, Mirol PM, Lirón JP, Dulout FN (2000). Male-mediated introgression of Bos indicus genes into Argentine and Bolivian Creole cattle breeds. Anim Genet 31: 302–305.
Giovambattista G, Ripoli MV, Peral García P, Bouzat JL (2001). Indigenous domestic breeds as reservoirs of genetic diversity: the Argentinean Creole cattle. Anim Genet 32: 240–247.
Hanotte O, Bradley DG, Ochieng JW, Verjee Y, Hill EW, Rege EO (2002). African pastoralism: genetic imprints of origins and migrations. Science 296: 336–339.
Higgins DG, Bleasby AJ, Funchs R (1992). CLUSTAL V: improved software for multiple sequence alignement. Comput Appl Biosci 8: 189–191.
Johnson HB (1987). Portuguese settlement, 1500–1580. In: Bethell L. (ed) Colonial Brazil, Cambridge University Press: Cambridge, UK. pp 1–38.
Kimura M (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120.
Lirón JP, Ripoli MV, Cancela L, De Luca JC, Dulout FN, Giovambattista G (2000). Genetic characterization of Creole cattle from Argentina, Bolivia and Uruguay. Anim Genet 31 (Suppl 1): 54.
Loftus RT, Ertugrul O, Harba AH, El-Barody MAA, MacHugh DE, Park SDE et al (1999). A microsatellite survey of cattle from a centre of origin: the Near East. Mol Ecol 8: 2015–2022.
Loftus RT, MacHugh DE, Bradley DG et al (1994a). Evidence for two independent domestications of cattle. Proc Natl Acad Sci USA 91: 2757–2761.
Loftus RT, MacHugh DE, Ngere LO, Balain DS, Badi AM, Bradley DG et al (1994b). Mitochondrial genetic variation in European, African and Indian cattle populations. Anim Genet 25: 265–271.
MacHugh DE, Shriver MD, Loftus RT, Cunningham EP, Bradley DG (1997). Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus). Genetics 146: 1071–1086.
Martin-Burriel I, García-Muro E, Zaragoza P (1999). Genetic diversity analysis of six Spanish native cattle breeds using microsatellites. Anim Genet 30: 177–182.
Meirelles FV, Rosa AJM, Lôbo RB, Garcia JM, Smith LC, Duarte FAM (1999). Is the American Zebu really Bos indicus? Genet Mol Biol 22: 543–546.
Moreno-Millán M, Rodero Franganillo A, Alonso Trujillo FJ (1991). Cytogenetic studies of retinta breed cattle: incidence of the 1/29 translocation. Inf Técn Econ Agrar 87A: 263–267.
Parry JH (1970). El imperio Español de Ultramar. Aguilar: Madrid.
Primo AT (1992). El ganado bovino ibérico en las Américas 500 años después. Arch Zootec 41: 421–432.
Rodero A, Delgado JV, Rodero E (1992). Primitive Andalusian livestock and their implications in the discovery of America. Arch Zootec 41: 383–400.
Sanchez-Belda A (1984). Razas bovinas españolas. Publicaciones de Extensión Agraria: Madrid, Spain.
Schneider S, Roessli D, Excoffier L (1997) . Arlequin: A Software for Population Genetics Data Analysis Ver 2.000. University of Geneva: Geneva, Switzerland.
Swofford DL (1997). paup: Phylogenetic Analysis Using Parsimony. Smithsonian Institution: Washington DC.
Troy CS, MacHugh DE, Bailey JF, Magee DA, Loftus RT, Cunningham P et al (2001). Genetic evidence for Near-Eastern origins of European cattle. Nature 410: 1088–1091.
Acknowledgements
This paper was supported by SECyT and Fundación Antorchas grants to PMM and GG, and National Research Council and Universidad Nacional de La Plata grants to FND. We thank Julio De Luca for the collection of samples and Chris Faulkes, Tamsin Burland and Steve Le Combe for very useful comments. The paper is dedicated to the memory of Ruth Vega.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mirol, P., Giovambattista, G., Lirón, J. et al. African and European mitochondrial haplotypes in South American Creole cattle. Heredity 91, 248–254 (2003). https://doi.org/10.1038/sj.hdy.6800312
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800312
Keywords
This article is cited by
-
Ancient DNA confirms diverse origins of early post-Columbian cattle in the Americas
Scientific Reports (2023)
-
The genetic ancestry of American Creole cattle inferred from uniparental and autosomal genetic markers
Scientific Reports (2019)
-
Resurrecting Darwin’s Niata - anatomical, biomechanical, genetic, and morphometric studies of morphological novelty in cattle
Scientific Reports (2018)
-
Variability of hair coat and skin traits as related to adaptation in Criollo Limonero cattle
Tropical Animal Health and Production (2011)