Abstract
We present the first clinical report of sibs with the multiple maternal hypomethylation syndrome. Both sisters presented with transient neonatal diabetes mellitus (TNDM). By methylation-specific PCR of bisulphite-treated DNA, we found a mosaic spectrum of hypomethylation at the following maternally methylated loci in both sibs: ZAC (6q24), KCNQ1OT1 (11p15.5), GRB10 (7p11.2–12), PEG3 (19q13), PEG1/MEST (7q32), and NESPAS (20q13). While the older sister has a milder phenotype, the younger one was severely ill and died at 11 months of age. Despite phenotypic differences, the sisters had several manifestations of both TNDM and BWS in common. The family is highly consanguineous, and the parents are first cousins. We suggest that the genetic defect in this family is a novel, most likely autosomal recessive defect of methylation mechanisms, either in the sisters or in their mother, affecting her oocyte imprinting. The recurrence with affected sibs as reported in this family has implications for genetic counselling.
Similar content being viewed by others
Introduction
Methylation of CpG dinucleotides in DNA is one of the epigenetic imprinting mechanisms through which alleles are differentially expressed, depending on their parental origin. We previously documented that aberrant methylation in imprinting disorders may affect more than one single locus: we found concurrent loss of methylation (LOM) of the imprinted transient neonatal diabetes mellitus (TNDM; MIM 601410) differentially methylated region (DMR) at 6q24 and Beckwith–Wiedemann syndrome (BWS; MIM 130650) DMR2 (KCNQ1OT1) at 11p15.5 in two TNDM patients.1 Similar dual LOM had previously been demonstrated in two patients by Arima et al.2 Subsequently, we showed that 6 out of 12 anonymised TNDM patients with LOM at TNDM DMR (6q24) in addition had a diverse spectrum of maternal hypomethylation at multiple imprinted loci including KCNQ1OT1 (11p15.5), and we suggested the existence of a maternal hypomethylation syndrome.3 A similar spectrum of multiple maternal hypomethylation was found by Rossignol et al,4 in 10 out of 40 LOM BWS patients.
With informed consent, we tested the methylation status at multiple imprinted loci in one of our two initial patients reported by Mackay et al1 and her younger sister who also had TNDM. We here describe their family history, clinical phenotypes, development, and laboratory results.
Methods
Clinical investigations
The family history was recorded from the mother, who gave her written informed consent. The medical records were reviewed in detail, and both girls were re-examined as part of the study.
Laboratory investigations
Methylation-specific PCR and pyrosequencing of bisulphite-induced polymorphisms
Methylation ratiometry of DNA from the sisters and their parents was performed for the loci ZAC (6q24), GRB10 (7p11.2–12), PEG1/MEST (7q32), KCNQ1OT1 and H19 (both 11p15.5), DLK1 (14q32), SNRPN (15q11–12), PEG3 (19q13), and NESPAS (20q13) by methylation-specific PCR (MS-PCR) following bisulphite treatment, as previously described.1, 3, 5 Primer sequences for NESPAS were derived from Williamson et al.6 Aberrant results were confirmed by either pyrosequencing7 or MS-PCR with another primer set. Control data were drawn from previous publications,1, 3 with supplementary MS-PCR at some loci to obtain an epigenotype of all 120 anonymised normal controls at all the loci. For the sisters, the MS-PCR was performed on genomic DNA extracted from EDTA-stabilised whole blood, cells from repeated buccal swabs, and skin fibroblasts.
Southern blot analysis
At the KCNQ1OT1 locus, the results of MS-PCR were also confirmed by Southern blot analysis with the LIT1 probe (AA155639) after digestion of genomic DNA with BamHI and NotI.8
Test for uniparental disomy by polymorphic microsatellite markers
Analysis of genomic DNA from the sisters and parents with the following polymorphic microsatellite markers was carried out to test for uniparental disomy: D6S292, D6S314, D6S1704 (6q23–24), D11S TH, D11S1318, D11S4088, and D11S4146 (11p15.4–15.5) (http://genome.ucsc.edu/).
Methylation-sensitive multiple ligation-dependent probe amplification analysis
Genomic and methylation-sensitive MLPA analysis of 11p15.5, using SALSA MS-MLPA kit ME030 (MRC Holland), was performed on leukocyte-derived DNA, to test for deletion or duplication of genes on 11p15.5, and to confirm the hypomethylation of KCNQ1OT1 in the sisters.
Karyotyping and screening for heterochromatin instability
Chromosome analysis of PHA-stimulated lymphocytes from the sisters and their parents was performed after G- and Q-banding (standard protocols; 450–500 banding level). One hundred metaphases from the younger sister were screened for micronuclei and paracentromeric heterochromatin instability, particularly of chromosomes 1 and 16, to exclude the immunodeficiency, centromeric instability, and facial anomalies syndrome (ICF syndrome; MIM 242860).
Subtelomeric MLPA analysis
To rule out a cryptic unbalanced translocation in the sisters, subtelomeric MLPA analysis was carried out with the SALSA MLPA kits P036B and P070 (MRC Holland).
Sequencing for KCNJ11 mutations
DNA from the older sister was sequenced for mutations in KCNJ11 by Molecular Genetics Laboratory, Royal Devon and Exeter NHS Healthcare Trust (Exeter, UK).
Screening for homozygosity at candidate gene loci by single-nucleotide polymorphisms (SNPs) and polymorphic microsatellites, and sequencing of the NLRP7 gene
By GeneChip® Mapping 10K 2.0. SNP Array (Affymetrix) and flanking microsatellite markers, genomic DNA from the sisters and their parents was analysed to screen for homozygosity at the following candidate gene loci: DNMT1 (19p13.3–13.2), DNMT2 (10p15.1), DNMT3A (2p23), DNMT3B (20q11.2), DNMT3L (21q22.3), and NLRP7 (19q13.4). Exons and intron–exon boundaries of NLRP7 in the mother were sequenced using the Golden Path sequence (http://genome.ucsc.edu/) and primers designed by ExonPrimer (http://ihg.gsf.de/ihg/ExonPrimer.html).
Results
Clinical investigations
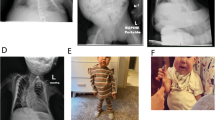
The sisters were the only children born to healthy consanguineous Turkish parents (Figure 1). The parents, maternal, and paternal grandparents are all first cousin couples. Five relatives have had either type 1 or 2 diabetes mellitus, or gestational diabetes. Several children in the family have died as neonates or in infancy for unknown reasons in Turkey. No relatives are known to have TNDM, BWS, or other imprinting disorders. A male cousin of the sisters suffers from epilepsy and psychomotor retardation of unknown cause.
The older sister1 has been followed up to the age of 5 years and 7 months (Figure 2); the younger sister until her death at 11 months of age. Their phenotypic features and developmental details are listed in Table 1.
Neonatally, the combination of hyperglycaemia, macroglossia, and intrauterine growth retardation in the older sister led to the clinical suspicion of TNDM. This diagnosis was subsequently confirmed by the finding of LOM at 6q24. At 7 months, the diabetes went into remission, but relapsed at the age of 2 years and 8 months. At 4 years and 6 months of age, insulin was successfully substituted with an oral sulphonyl urea, which is still her medication (tolbutamide, 500 mg three times a day). Her postnatal growth, in contrast to prenatal growth, is significantly accelerated, and her psychomotor development is mildly retarded.
The younger sister also presented with neonatal diabetes and, initially, seemed to be relatively mildly affected. From about 3 months of age, however, she had increasing failure to thrive and was repeatedly hospitalised due to recurrent infections. Clinically, she was suspected of an immunodeficiency, but immunoglobulins and Mannan-binding lectin were normal. She had severe growth retardation with no postnatal catch-up, and gross psychomotor retardation. She required insulin until she died.
A comparison between the clinical features of the sisters, and the features of TNDM and BWS is shown in Table 2.
Laboratory investigations
The results of the MS-PCR are shown in Table 3, and examples of the methylation analyses appear in Figure 3.
Examples of the methylation analyses performed. (a) MS-PCR analysis at KCNQ1OT1 in BWS DMR2. X-axis: product size (bp); Y-axis: peak height. The ratio T/C was calculated as the height ratio of unmeth/meth products, and normalised to the average of normal control samples. Both sisters have hypomethylation of the maternal allele in DNA from blood and normal methylation in DNA obtained by mouth brush. (b) Methylation-specific Southern blot analysis of KCNQ1OT1 (LIT1) in BWS DMR2. Restriction enzymes: BamHI and NotI. Probe: LIT1 (AA155639). Hy=hypomethylation; N=normal; O=older sister; Y=younger sister; M=mother; F=father. Both sisters have hypomethylation of the maternal allele in blood-derived DNA and normal methylation in DNA from fibroblasts. (c) MS-PCR analysis at PEG3. Method similar to that used in (a). Both sisters have complete loss of maternal methylation. (d) Quantitative pyrosequencing of bisulphate-induced C/T polymorphisms (shaded areas) at PEG3. Both sisters have strongly elevated T/C ratios, indicating marked maternal hypomethylation.
Both sisters had hypomethylation at multiple imprinted, maternally methylated loci. The hypomethylation was mosaic with respect to the cell types affected and level of methylation. At TNDM DMR (6q24) and GRB10 (7p11.2–12), both sisters had complete LOM in all cell types analysed, while there was a spectrum of hypomethylation at KCNQ1OT1 (11p15.5), PEG3 (19q13), PEG1/MEST (7q32), and NESPAS (the younger sister had only minimal alteration in methylation at NESPAS). SNRPN (15q11–12), H19 (BWS DMR1; 11p15.5), and the DLK1 DMR (14q32) showed normal methylation. No hypomethylation was detected in any control samples. The parents had normal methylation at all loci.
The results of methylation-sensitive multiple ligation-dependent probe amplification (MS-MLPA) analysis of 11p15.5 on blood-derived DNA indicated a moderate mosaic hypomethylation at KCNQ1OT1 in the older sister, confirming the results of MS-PCR and Southern blot (data not shown). For the younger sister, the result of MS-MLPA at this locus was within normal limits, which is in accordance with our finding that subtle methylation abnormalities may go undetected by this method (personal observation). MS-MLPA showed no evidence of allele copy number change in the sisters.
Polymorphic microsatellite marker analysis showed normal, biparental inheritance, and no evidence of deletion or duplication of chromosomes 6q23–24 and 11p15.5.
The sisters and their parents all had normal karyotypes of lymphocytes, and there were no micronuclei or heterochromatin instability, indicating ICF syndrome in the younger sister.
Subtelomeric MLPA analyses were normal, thus excluding unbalanced translocations in the sisters.
No KCNJ11 mutation was found in the older sister at sequencing.
Analysis at the DNMT genes and NLRP7, of SNPs and polymorphic microsatellites in combination showed heterozygosity in one or both sisters, and the sisters had inherited divergent parental alleles around these loci. At SNP analysis, the mother was heterozygous at all DNMT loci, except for DNMT1, where she, like her husband and daughters, was homozygous for 8.2 Mb around the gene. When analysed with four polymorphic microsatellite markers spanning 1.58 Mb around DNMT1, she was, however, heterozygous for three of them. We conclude that the homozygosity of SNPs around DNMT1 was due to non-informativity. The mother was heterozygous around NLRP7, and sequencing her NLRP7 revealed no mutations.
Discussion
This is the first clinical report of sibs affected with hypomethylation at multiple imprinted, maternally methylated loci. The underlying mechanism of this novel syndrome is not known. Environmental factors may play a role in the complex processes of establishing and maintaining methylation imprinting marks in both animals9 and humans, for example, related to assisted reproductive technology.10 In the family presented here with affected sibs born after naturally conceived pregnancies, the aetiology of the multiple methylation aberrations is likely to be genetic. The imprinted loci found to be affected in this study are all maternally methylated in normal individuals, while the paternally methylated loci had normal methylation. Furthermore, hypomethylation was mosaic with respect to its level, the cell types and tissues affected, and the loci involved, suggesting a postzygotic origin of the aberrant pattern of DNA methylation.
Phenotypic findings
Despite many differences, the clinical phenotypes of the sisters had several features in common including a presentation with intrauterine growth retardation, neonatal diabetes, macroglossia, and umbilical defects (see Table 1). Complete LOM at TNDM DMR in both sibs could explain these findings. Macroglossia and abdominal wall defect as well as the postnatal excessive growth in sib 1 could, however, also result from the hypomethylation at KCNQ1OT1, as these manifestations are major features of BWS (Table 2). Postaxial polydactyly (sib 1) and cardiomyopathy (sib 2) can also occasionally occur in BWS patients.11, 12 Sib 1's relapse of diabetes at 2 years and 8 months was early, compared to previously reported TNDM cases, where the average was 14 years with an earliest reported age of 4 years prior to this report.13 It is possible that the obesity of the patient provoked an insulin resistance and thus an early relapse of the diabetes. We do not know whether the diabetes of the younger sister was transient or permanent; she still required insulin at the time of her death when she was 11 months old, but her diabetes may have been prolonged by the recurrent severe infections and cardiac failure.
Loss of maternal-specific methylation at NESPAS, part of the complex, imprinted GNAS cluster on chromosome 20q13, is also associated with a well-recognised clinical phenotype; pseudohypoparathyroidism type IB (PHP IB; MIM 603233).14 However, features of this condition such as hypocalcaemia, hyperphosphataemia, and elevated parathyroid hormone were not seen in either sib (Table 1). We interpret the slight TSH elevation with normal T3 and T4 in the older sister on one occasion as an incidental finding. Neither sib had any features suggestive of Albright hereditary osteodystrophy (MIM 103580). The clinical consequences of hypomethylation in humans at GRB10, PEG3, and PEG1/MEST are not known and may account for some of the unexplained findings described in Table 1. Alternatively, these may result from methylation aberrations at loci or tissues not tested.
There are significant differences in the clinical phenotypes of the sisters; most notably, the repeated infections, epilepsy, severe psychomotor retardation, failure to thrive, and early death of the younger sister. These phenotypic differences may reflect differences in the methylation patterns in the two sibs. Alternatively, given the high degree of consanguinity, it is possible that the severity of symptoms in the younger sister was the result of a second genetic disorder.
As regards the differential diagnosis, we considered two rare genetic conditions with methylation aberrations at multiple sites. The ICF syndrome is caused by mutations in the DNMT3B gene,15 affecting the methylation and condensation of paracentromeric heterochromatin; micronuclei and centromeric instability of chromosomes 1 and 16, and, less often, 9 are typically found. ICF patients have variable types of immunodeficiency and a characteristic dysmorphic appearance with hypertelorism, protruding tongue, and micrognathia.16 While the severe infections seen in sib 2 and macroglossia are somewhat reminiscent, the other clinical findings seen in this family are different, and no centromeric instability or abnormalities of immunoglobulins were demonstrated.
Aberrant maternal methylation at multiple imprinted loci has been demonstrated in familial recurrent biparental complete hydatidiform mole (HYDM; MIM 231090).17, 18 This is also autosomal recessively inherited. In the majority of cases, biparental complete HYDM is caused by mutations in the NLRP7 gene (HNGC approved nomenclature; synonym NALP7).19 Homozygosity in affected women causes a dysregulation of imprinting in the germ line and leads to successive moles or occasionally other reproductive losses. This represents a global maternal imprinting defect incompatible with fetal life.20 Allelic mutations in NLRP7 could, however, hypothetically account for the multiple maternal hypomethylation syndrome, pointing to the possibility that the gene defect in this syndrome may affect either the sibs or their mother. It must be noted that there was no history of recurrent moles or pregnancy losses in the family. Microsatellite markers and SNPs around NLRP7 showed that the mother was heterozygous, and furthermore, sequencing her NLRP7 exons and intron–exon boundaries revealed no mutations. That is, a NLRP7 mutation in the mother is unlikely as the underlying defect in this family, although it is not completely ruled out. Other potential candidate genes are the DNA methyltransferases involved in the establishment or maintenance of DNA methylation: DNMT1 and DNMT1o (its oocyte-specific isoform), DNMT2, DNMT3A, DNMT3B, and DNMT3L. Our analysis of SNPs and microsatellites did not point specifically to any one of these genes as a candidate for a homozygous mutation in the mother. In mice, however, embryos derived from Dnmt1o-deficient oocytes have been found to have loss of allele-specific expression and methylation, specifically at imprinted loci.21 Heterozygous progeny of female mice null mutant for Dnmt3L have been shown to have specific demethylation of maternally methylated, imprinted genes.22 Therefore, based on these mice data, both of these genes could be candidates, and further studies are in progress.
Clinical implications
The clinical findings in the older sister initially pointed to a diagnosis of TNDM (Table 2), which was subsequently confirmed by genetic testing. Therefore, the diagnosis of hypomethylation at other loci such as KCNQ1OT1 was substantially delayed, although a suspicion of BWS was raised from birth. On the basis of this finding, we recommend that TNDM patients who are primarily diagnosed with LOM at 6q24, should also be tested for LOM at 11p15.5, so that if a molecular diagnosis of BWS is made, surveillance for embryonal tumours can be offered.
The risk of recurrence of this novel syndrome is not known. Initially, when only the TNDM DMR at 6q24 was known to be involved, the mother was counselled that the recurrence risk of LOM TNDM was likely to be low, as all cases so far had been sporadic, however, with the caution that LOM TNDM is a very rare condition. The birth of the second affected child and the detection of hypomethylation at further loci make it likely that the disorder in this family is genetic with a significant reproductive risk. However, the exact risk to future offspring is difficult to know. If the syndrome is caused by a homozygous autosomal recessive defect in the sisters, the risk is 25%. But if instead the mother is homozyogous for such a defect affecting the imprinting of her oocytes, the situation is comparable to biparental complete HYDM, where affected women have a theoretical recurrence risk of 100%. Selective mechanisms, however, modify this risk figure in biparental complete HYDM, as in a few families, some of the affected women have experienced normal pregnancies.20 Finally, due to the very broad clinical spectrum in the sisters, it is currently impossible to predict the phenotype associated with multiple hypomethylation. Further knowledge about the underlying mechanisms and the associated phenotypic spectrum in multiple maternal hypomethylation syndrome is needed.
References
Mackay DJG, Hahnemann JMD, Boonen SE et al: Epimutation of the TNDM locus and the Beckwith–Wiedemann syndrome centromeric locus in individuals with transient neonatal diabetes mellitus. Hum Genet 2006; 119: 179–184.
Arima T, Kamikihara T, Hayashida T et al: ZAC, LIT1(KCNQ1OT1) and p57KIP2(CDKN1C) are in an imprinted gene network that may play a role in Beckwith–Wiedemann syndrome. Nucleic Acids Res 2005; 33: 2650–2660.
Mackay DJG, Boonen SE, Clayton-Smith J et al: A maternal hypomethylation syndrome presenting as transient neonatal diabetes mellitus. Hum Genet 2006; 120: 262–269.
Rossignol S, Steunou V, Chalas C et al: The epigenetic imprinting defect of patients with Beckwith–Wiedemann syndrome born after assisted reproductive technology is not restricted to the 11p15 region. J Med Genet 2006; 43: 902–907.
Mackay DJG, Temple IK, Shield JPH, Robinson DO : Bisulphite sequencing of the transient neonatal diabetes mellitus DMR facilitates a novel diagnostic test but reveals no methylation anomalies in patients of unknown aetiology. Hum Genet 2005; 116: 255–261.
Williamson CM, Turner MD, Ball ST et al: Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet 2006; 38: 350–355.
Dupont JM, Tost J, Jammes H, Gut IG : De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal Biochem 2004; 333: 119–127.
Mitsuya K, Meguro M, Lee MP et al: LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum Mol Genet 1999; 8: 1209–1217.
Richards EJ : Inherited epigenetic variation – revisiting soft inheritance. Nat Rev Genet 2006; 7: 395–401.
Horsthemke B, Ludwig M : Assisted reproduction: the epigenetic perspective. Hum Reprod Update 2005; 11: 473–482.
Weksberg R, Shuman C : Beckwith–Wiedemann syndrome and hemihyperplasia; in Cassidy SB, Allanson JE (eds): Management of Genetic Syndromes. Wiley-Liss, Inc.: New York, 2005, 2nd edn, pp 101–115.
Elliott M, Bayly R, Cole T, Temple IK, Maher ER : Clinical features and natural history of Beckwith–Wiedemann syndrome: presentation of 74 new cases. Clin Genet 1994; 46: 168–174.
Temple IK, Shield JP : Transient neonatal diabetes, a disorder of imprinting. J Med Genet 2002; 39: 872–875.
Liu J, Nealon JG, Weinstein LS : Distinct patterns of abnormal GNAS imprinting in familial and sporadic pseudohypoparathyroidism type IB. Hum Mol Genet 2005; 14: 95–102.
Xu GL, Bestor TH, Bourc'his D et al: Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 1999; 402: 187–191.
Ehrlich M, Jackson K, Weemaes C : Immunodeficiency, centromeric region instability, facial anomalies syndrome (ICF). Orphanet J Rare Dis 2006; 1: 2.
Judson H, Hayward BE, Sheridan E, Bonthron DT : A global disorder of imprinting in the human female germ line. Nature 2002; 416: 539–542.
El-Maarri O, Seoud M, Coullin P et al: Maternal alleles acquiring paternal methylation patterns in biparental complete hydatidiform moles. Hum Mol Genet 2003; 12: 1405–1413.
Murdoch S, Djuric U, Mazhar B et al: Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet 2006; 38: 300–302.
Van den Veyver IB, Al-Hussaini TK : Biparental hydatidiform moles: a maternal effect mutation affecting imprinting in the offspring. Hum Reprod Update 2006; 12: 233–242.
Howell CY, Bestor TH, Ding F et al: Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 2001; 104: 829–838.
Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH : Dnmt3L and the establishment of maternal genomic imprints. Science 2001; 294: 2536–2539.
Acknowledgements
We thank the family for their collaboration. We thank the technicians at the Kennedy Center and Jonathan Callaway, Salisbury, for their laboratory work and photographer Jette B Rasmussen for excellent assistance with preparation of the figures. Deborah JG Mackay was funded by Diabetes UK.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Boonen, S., Pörksen, S., Mackay, D. et al. Clinical characterisation of the multiple maternal hypomethylation syndrome in siblings. Eur J Hum Genet 16, 453–461 (2008). https://doi.org/10.1038/sj.ejhg.5201993
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201993
Keywords
This article is cited by
-
Transient neonatal diabetes mellitus and hypomethylation at additional imprinted loci: novel ZFP57 mutation and review on the literature
Acta Diabetologica (2019)
-
Beckwith–Wiedemann syndrome and pseudohypoparathyroidism type Ib in a patient with multilocus imprinting disturbance: a female-dominant phenomenon?
Journal of Human Genetics (2016)
-
Epigenetic and genetic alterations of the imprinting disorder Beckwith–Wiedemann syndrome and related disorders
Journal of Human Genetics (2013)
-
Transienter neonataler Diabetes und Hypomethylierungssyndrome
Medizinische Genetik (2010)
-
6q24 transient neonatal diabetes
Reviews in Endocrine and Metabolic Disorders (2010)