Abstract
Beckwith–Wiedemann syndrome (BWS) is an imprinting-related human disease. The frequencies of causative alterations such as loss of methylation (LOM) of KvDMR1, hypermethylation of H19-DMR, paternal uniparental disomy, CDKN1C gene mutation, and chromosome abnormality have been described for North American and European patients, but the corresponding frequencies in Japanese patients have not been measured to date. Analysis of 47 Japanese cases of BWS revealed a significantly lower frequency of H19-DMR hypermethylation and a higher frequency of chromosome abnormality than in North American and European patients. These results suggest that susceptibility to epigenetic and genetic alterations differs between the two groups.
Similar content being viewed by others
Introduction
Beckwith–Wiedemann syndrome (BWS), a well-known imprinting-related human disease, is characterized by macrosomia, macroglossia, and abdominal wall defects (OMIM #130650). Genomic imprinting is an epigenetic phenomenon that is responsible for parent-of-origin-specific expression of genes. The relevant imprinted chromosomal region in BWS, 11p15.5, consists of two independent imprinted domains, IGF2/H19 and CDKN1C/LIT1. Imprinted genes within each domain are regulated by the imprinting control region (ICR), which in this case is either H19-DMR or KvDM1 (Figure 1a).1 In North American and European BWS patients, several causative alterations have been identified: KvDMR1 loss of methylation (LOM) (∼50%), H19-DMR hypermethylation (2–7%), paternal uniparental disomy (patUPD; ∼20%), CDKN1C mutations (∼10%), duplications of 11p15 (<1%), and inversions or translocations involving 11p15 (<1%).1 The cause is unknown for ∼15% of patients.1 Comprehensive analyses of Japanese patients with BWS, however, have not been carried out to date. To add to our understanding of frequencies of epigenetic mutations in the Japanese population, we analyzed 47 cases of BWS. Compared with North American and European groups, the frequency of H19-DMR hypermethylation was significantly lower (0%) and that of chromosome abnormality (13%) was higher in the Japanese patients. These results suggest that susceptibility to epigenetic and genetic alterations differs according to ethnicity.
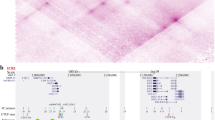
11p15.5 imprinted region and methylation and patUPD analysis. (a) 11p15.5 imprinted region. Representative genes are shown. Gray boxes and shaded boxes indicate maternal and paternal expressed genes, respectively. KvDMR1 and H19-DMR are the ICRs for each domain. Black and white ovals indicate methylated and unmethylated ICRs, respectively. Broken arrows indicate the transcriptional direction. (b, c) Representative results for methylation analyses of KvDMR1 and H19-DMR using hot-stop COBRA. patUPD patients had partial KvDMR1 LOM and partial H19-DMR hypermethylation. (d) Representative results for patUPD analysis. Bands transmitted from the father were stronger than those from the mother, indicating patUPD. 18P and 19P, DNA from patient's father; 18M and 19M, DNA from patient's mother; 18S, unaffected sister; c, control for complete digestion. MC, methylated DNA control; NC, normal control.
Materials and methods
BWS patients
Forty-seven Japanese patients who had been diagnosed with BWS by clinical geneticists were analyzed. Diagnoses were made according to the criteria of DeBaun and Tucker2 with all patients having at least two of the five most common features (macroglossia, birth weight >90th percentile, hypoglycemia in the first month of life, ear creases or ear pits, and abdominal wall defects). DNA was extracted from lymphoblastoid cell lines, peripheral blood lymphocytes, or tissues. One sample from each patient was used. This study was approved by the Ethical Committee for Human Genome and Gene Analyses of the Faculty of Medicine, Saga University.
Chromosomal analysis
Metaphase chromosomes were analyzed using the standard G-banding method.
Methylation analyses
We investigated methylation status at the AccII site 16 bp downstream of the NotI site in KvDMR1, and the MluI site approximately 80 bp downstream of the CTCF binding site 6 (CTS6) in H19-DMR. Combined bisulfite restriction analyses (COBRA) using the hot-stop method were employed as described previously.3, 4 For H19-DMR, bisulfite sequencing was performed using the same primers as used for COBRA. PCR primers and conditions are shown in Supplementary Table 1. The percentage methylation was calculated as: intensity of methylated band/(intensity of methylated band+intensity of unmethylated band) × 100.
Paternal uniparental disomy
To analyze patUPD, we used DNA polymorphic markers for 11p15.5, including tetranucleotide repeats in the tyrosine hydroxylase gene (TH), an AvaII (ApaI) polymorphism in IGF2, an RsaI polymorphism in H19, and an NsiI polymorphism in KCNQ1OT1. Hot-stop PCR was employed for PCR–RFLP to eliminate heteroduplex formation. The percentage mosaicism of patUPD was calculated as:5 % mosaicism=(k−1)/(k+1) × 100, where k is the ratio of the intensity of the paternal to maternal alleles of the sample. We defined patUPD as more than 30% mosaicism.
CDKN1C mutations
Mutations of CDKN1C were investigated by sequencing as described previously.6
Statistical analysis
Differences between Japanese and North American and European patients with BWS were calculated using the χ2-test or Fisher's direct probability method. Probability levels of <0.05 were considered statistically significant.
Results and discussion
We used hot-stop COBRA for quantitative analysis of methylation, the results of which were confirmed in several patients by methylation-sensitive Southern blot analysis. We found that 15 patients (#1–15) had complete LOM of KvDMR1 and 7 patients (#16–18, #20–22, #28) had partial LOM (Table 1 and Figure 1b). We also found seven patients (#16–19, #27–29) with partial hypermethylation of H19-DMR, but found no patient with complete hypermethylation (Figure 1c). patUPD manifested in a mosaic pattern, and trisomy 11 was associated with an extra 11p, which may have affected the results of methylation analyses at KvDMR1 and H19-DMR. Indeed, all our patients with partial KvDMR1 LOM and/or partial H19-DMR hypermethylation had mosaic patUPD (#16–22) or trisomy 11 (#27–29; Figure 1b and c). We thus concluded that 15 (#1–15) patients (34%) had isolated KvDMR1 LOM. For H19-DMR, since the MluI site used for COBRA was approximately 80 bp downstream of CTS6, we examined whether the methylation status of the MluI site correctly reflected that within the core sequence of CTS6 by bisulfite sequencing. A total of 258 clones generated from 13 patients (11–20 clones per patient; 5 patients with KvDMR1 LOM, 3 with patUPD, 1 with the KIP2 mutation, and 4 with cause unknown) and 5 normal individuals (11–12 clones per individual) were sequenced. We found that 235 clones (91%) showed the same methylation status at the MluI site and CTS6 (data not shown). Thus, we concluded that COBRA based on the MluI site accurately reflected the methylation status of CTS6. We also concluded that no patient in this study had isolated H19-DMR hypermethylation.
When PCR products generated from the paternal allele were more intense than those from the maternal allele, an allelic imbalance was indicated (Figure 1d). We found seven patients with patUPD (15%), as mentioned above, and six patients (#27–32) with chromosome abnormalities (13%); via karyotyping, four of these patients were found to have translocations and two were found to have trisomy 11 (Table 1). Two (#27 and #28) of the four translocation patients had partial trisomy for 11p due to a translocation. Partial KvDMR1 LOM and/or partial H19-DMR hypermethylation in patients #27–29 indicated that they had two copies of paternal 11p. Patient #27 was previously described as having paternal origin duplication of 11p.7 Whether the chromosomal abnormalities in the remaining two cases (#30 and #31) were of paternal origin was unknown. Although patient #32, who suffered from umbilical hernia, macroglossia, ear creases, and ear pits, had a translocation, it did not involve 11p or any molecular abnormalities. Thus, an unknown causative factor, which may or may not be due to the translocation, is involved in this patient. We found three mutations in CDKN1C, that is, 399C>T, 570delCTinsG, and 1086delTinsAG, in four patients (9%; #23–#26; #24 and #25 were siblings), as reported previously.8
Of the 47 patients, 15 (32%; #33–47) did not have any identifiable alterations, and so their BWS was designated as cause unknown. The frequencies of macrosomia and abdominal wall defects were lower in these patients than in patients with known causes (data not shown). Although microdeletion of H19-DMR has been reported,9, 10, 11 we did not find it in any patient examined, including the 15 patients with cause unknown and two patients (#18 and #28) who had patUPD and trisomy 11 due to t(11;X), respectively (data not shown).
The frequency of isolated H19-DMR hypermethylation was significantly lower in Japanese patients with BWS than in the North American and European patients (Table 2). The Japanese patients had more frequent chromosomal abnormality. Although our sample number was not large, our data suggest that susceptibility to epigenetic and genetic alterations leading to BWS varies according to ethnicity. BWS predisposes patients to embryonal tumors, especially Wilms’ tumor, in which loss of imprinting (LOI) of IGF2 accompanied by H19-DMR hypermethylation is involved. Furthermore, a strong association between H19-DMR hypermethylation and Wilms’ tumor development in BWS has been reported.12, 13 In Japanese BWS patients, however, the frequency of Wilms’ tumor tends to be lower than expected,14 and low frequencies of H19-DMR hypermethylation and IGF2 LOI in Wilms’ tumors in Japanese children have been reported.15 Of the 47 patients in the present study, 5 patients developed tumors (3 hepatoblastomas, 1 rhabdomyosarcoma, and 1 cardiac atrial tumor). Although overall tumor incidence (11%) was consistent with that reported for North American and European patients, there were no patients with Wilms’ tumor in our sample. The low frequency of H19-DMR hypermethylation may account for this. Further investigations will be necessary to understand whether the different frequencies of epigenetic and genetic alterations are due to DNA polymorphisms, such as SNPs and/or low copy repeats, at 11p.
References
Weksberg R, Shuman C, Smith AC : Beckwith–Wiedemann syndrome. Am J Med Genet C Semin Med Genet 2005; 137: 12–23.
DeBaun MR, Tucker MA : Risk of cancer during the first four years of life in children from the Beckwith–Wiedemann Syndrome Registry. J Pediatr 1998; 132: 398–400.
Soejima H, Nakagawachi T, Zhao W et al: Silencing of imprinted CDKN1C gene expression is associated with loss of CpG and histone H3 lysine 9 methylation at DMR-LIT1 in esophageal cancer. Oncogene 2004; 23: 4380–4388.
Satoh Y, Nakadate H, Nakagawachi T et al: Genetic and epigenetic alterations on the short arm of chromosome 11 are involved in a majority of sporadic Wilms’ tumours. Br J Cancer 2006; 95: 541–547.
Itoh N, Becroft DM, Reeve AE, Morison IM : Proportion of cells with paternal 11p15 uniparental disomy correlates with organ enlargement in Wiedemann–Beckwith syndrome. Am J Med Genet 2000; 92: 111–116.
Hatada I, Ohashi H, Fukushima Y et al: An imprinted gene p57KIP2 is mutated in Beckwith–Wiedemann syndrome. Nat Genet 1996; 14: 171–173.
Mannens M, Hoovers JM, Redeker E et al: Parental imprinting of human chromosome region 11p15.3-pter involved in the Beckwith–Wiedemann syndrome and various human neoplasia. Eur J Hum Genet 1994; 2: 3–23.
Hatada I, Nabetani A, Morisaki H et al: New p57KIP2 mutations in Beckwith–Wiedemann syndrome. Hum Genet 1997; 100: 681–683.
Sparago A, Cerrato F, Vernucci M, Ferrero GB, Silengo MC, Riccio A : Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith–Wiedemann syndrome. Nat Genet 2004; 36: 958–960.
Prawitt D, Enklaar T, Gartner-Rupprecht B et al: Microdeletion of target sites for insulator protein CTCF in a chromosome 11p15 imprinting center in Beckwith–Wiedemann syndrome and Wilms’ tumor. Proc Natl Acad Sci USA 2005; 102: 4085–4090.
Sparago A, Russo S, Cerrato F et al: Mechanisms causing imprinting defects in familial Beckwith–Wiedemann syndrome with Wilms’ tumour. Hum Mol Genet 2007; 16: 254–264.
Weksberg R, Nishikawa J, Caluseriu O et al: Tumor development in the Beckwith–Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum Mol Genet 2001; 10: 2989–3000.
Rump P, Zeegers MP, van Essen AJ : Tumor risk in Beckwith–Wiedemann syndrome: a review and meta-analysis. Am J Med Genet A 2005; 136: 95–104.
Kobayashi Y : Malformation syndromes and childhood tumors; in Shimizu K, Misugi K (eds): Shouni Geka Byourigaku (Pediatric Surgical Pathology). Tokyo: Bunkodo, 1995, pp 12–18.
Fukuzawa R, Breslow NE, Morison IM et al: Epigenetic differences between Wilms’ tumours in white and east-Asian children. Lancet 2004; 363: 446–451.
Gaston V, Le Bouc Y, Soupre V et al: Analysis of the methylation status of the KCNQ1OT and H19 genes in leukocyte DNA for the diagnosis and prognosis of Beckwith–Wiedemann syndrome. Eur J Hum Genet 2001; 9: 409–418.
DeBaun MR, Niemitz EL, McNeil DE, Brandenburg SA, Lee MP, Feinberg AP : Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith–Wiedemann syndrome with cancer and birth defects. Am J Hum Genet 2002; 70: 604–611.
Bliek J, Gicquel C, Maas S, Gaston V, Le Bouc Y, Mannens M : Epigenotyping as a tool for the prediction of tumor risk and tumor type in patients with Beckwith–Wiedemann syndrome (BWS). J Pediatr 2004; 145: 796–799.
Acknowledgements
We express our sincere gratitude to the patients and their families. We also thank Drs Makoto Takeuchi (Osaka Medical Center and Research Institute for Maternal and Child Health), Mayumi Iwakawa (National Institute of Radiological Sciences), Tateo Kuno and Yuhei Hamasaki (Saga University Faculty of Medicine), Akira Shimada (Gunma Children’s Medical Center), Hidefumi Tonoki (Tenshi Hospital), Noboru Takizawa (Graduate School of Medical Sciences, Kanazawa University), Chiga Tokuda (Nantan General Hospital), Tsuyako Iwai (Kagawa Children's Hospital), Asami Sato and Takanori Yamagata (Jichi Medical School), Ikuko Takahashi (Akita University), Toshiro Nagai (Koshigaya Hospital, Dokkyo Medical University), Akira Yoshioka (Nara Medical University), and Kenji Ihara (Kyushu University Faculty of Medicine) for providing and caring for the patients. This study was supported in part by a Grant-in-Aid for Scientific Research (C) (No. 18590313) from the Japan Society for the Promotion of Science and by Grants-in-Aid for the Third Term Comprehensive Ten-Year Strategy for Cancer Control from the Ministry of Health, Labour and Welfare of Japan.
Author information
Authors and Affiliations
Additional information
Competing interests
None declared.
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Sasaki, K., Soejima, H., Higashimoto, K. et al. Japanese and North American/European patients with Beckwith–Wiedemann syndrome have different frequencies of some epigenetic and genetic alterations. Eur J Hum Genet 15, 1205–1210 (2007). https://doi.org/10.1038/sj.ejhg.5201912
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201912
Keywords
This article is cited by
-
Association of four imprinting disorders and ART
Clinical Epigenetics (2019)
-
Placenta-specific epimutation at H19-DMR among common pregnancy complications: its frequency and effect on the expression patterns of H19 and IGF2
Clinical Epigenetics (2019)
-
De novo paternal origin duplication of chromosome 11p15.5: report of two Chinese cases with Beckwith-Wiedemann syndrome
Molecular Cytogenetics (2017)
-
A high incidence of WT1 abnormality in bilateral Wilms tumours in Japan, and the penetrance rates in children with WT1 germline mutation
British Journal of Cancer (2015)
-
Epigenetic and genetic alterations of the imprinting disorder Beckwith–Wiedemann syndrome and related disorders
Journal of Human Genetics (2013)