Abstract
Metanephric adenoma has morphologic similarities to papillary renal cell neoplasms. Cytogenetic studies of papillary renal cell carcinoma and papillary adenoma have shown frequent gains of chromosomes 7 and 17 and loss of the Y chromosome. Some cytogenetic studies have supported the hypothesis that metanephric adenoma is related to papillary renal cell neoplasia; others have not. Seven metanephric adenomas were studied with fluorescence in situ hybridization in paraffin sections using centromeric probes for chromosomes 7, 17, and Y diluted 1:100 with tDenHyb1 buffer. The signals in 100 to 200 nuclei were counted in each tumor. Samples of histologically normal renal cortical tubule epithelium were used as controls. In all seven metanephric adenomas, the results for chromosomes 7 and 17 were similar: a high percentage of nuclei with two signals (range, 75 to 85%; median, 79%). Normal kidney showed similar results (range, 78 to 88%; median, 84%). The Y chromosome was present in all three of the tumors from males (range, 86 to 89% of nuclei; median, 87%). Normal kidney gave similar results (range 82% to 91%, median 84%). The presence of chromosomes 7, 17, and Y in metanephric adenomas is similar to their presence in normal kidney. Metanephric adenoma lacks the frequent gains of chromosomes 7 and 17 and losses of the Y chromosome that are typical of papillary renal cell neoplasms, supporting the notion that metanephric adenoma is not related to papillary renal cell carcinoma and papillary adenoma. Genetic analysis of chromosomes 7, 17, and Y may facilitate discrimination of metanephric adenoma from papillary renal cell carcinoma in difficult cases.
Similar content being viewed by others
INTRODUCTION
Metanephric adenoma is a rare benign neoplasm of the kidney, predominantly occurring in women and histologically similar to developing metanephric tubular epithelium (1, 2, 3). The histogenesis of metanephric adenoma and its relationship to intralobar nephrogenic rests, nephroblastoma, and papillary renal cell neoplasia is still a matter of controversy (2, 3, 4). However, current classifications recognize it as a distinct renal neoplasm (5, 6). The overlap in morphologic features between metanephric adenoma and papillary renal cell neoplasia has frequently been noted (2, 3, 4, 7). Immunohistochemistry and lectin histochemistry have given varied results in different laboratories and consequently have shed little light on its relationship to papillary renal cell neoplasms (8).
Cytogenetic studies of metanephric adenoma have been limited and have yielded conflicting results. In a study of 11 tumors, Brown et al. (9) found cytogenetic abnormalities similar to those typical of papillary renal cell carcinomas in 8 tumors. However, several reports of one or two tumors have found no gains of chromosomes 7 and 17, and no loss of Y (2, 10, 11, 12, 13, 14, 15). The present study was undertaken to determine the numbers of chromosomes 7, 17 and Y in seven metanephric adenomas, using fluorescence in situ hybridization with centromeric probes in paraffin-embedded tissues and to determine the similarities or differences with the pattern of gains and losses for these chromosomes, which is typical of papillary renal cell neoplasia (16, 17, 18).
MATERIALS AND METHODS
Tissue Samples
Seven metanephric adenomas were collected from personal consultation materials from one of the authors (JNE). The patients were four women (ages 29, 41, 43, and 49 y) and three men (ages 2, 27, and 53 y). With the exception of a 5-mm metanephric adenoma that was found incidentally in a kidney removed for angiomyolipoma, the metanephric adenomas ranged from 30 to 55 mm in greatest diameter. Twenty-seven histogically normal kidney samples were controls for chromosome 7, 61 histologically normal kidney samples for chromosome 17, and 18 samples from males for chromosome Y.
Fluorescence In Situ Hybridization
From each tumor, sections 5 μm thick were cut from paraffin-embedded blocks. The paraffin was removed from the sections with two 10-minute washes in xylene. After hydrating in 100%, 85%, and 70% ethanol solutions (10 min), rinsing in distilled water (10 min), and twice in phosphate buffer solution (pH 7, 10 min each), the slides were fixed in methanol-acetic acid 3:1 for 10 minutes and air dried. The sections were treated in a 2× standard saline citrate (SSC) solution for 15 minutes at 37° C and then dehydrated in consecutive 70%, 85%, and 100% ethanol solutions for one minute each and then dried. The sections were bathed in 0.1 mm citric acid (pH 6) solution at 85° C for 1 hour. Then they were again dehydrated in a series of ethanol solutions and dried. The tissue was digested by applying 0.75 mL of pepsin (Sigma, St. Louis, MO) solution (4 mg/mL in 0.9% NaCl, pH 1.5) to each slide and incubating them in a humidified box for 30 minutes at 37° C. Next, the slides were rinsed with distilled water for few seconds, dehydrated again in graded ethanol solutions and dried. Centromeric probes for chromosomes 7, 17, and Y (Vysis, Downers Grove, IL) were used. Each probe was diluted 1: 100 in tDenHyb1 buffer (Insitus, Albuquerque, NM). Ten μL of diluted probe was applied to each slide, and coverslips were placed over the slides. Denaturation was achieved by incubating the slides at 80° C for 10 minutes in a humidified box; then hybridization was done at 37° C for 3 hours. The coverslips were then removed and the slides were immersed at room temperature in 0.5× SSC for 2 minutes, in 50% formamide–1× SSC for 5 minutes, and in 2× SSC for 2 minutes. The slides were air dried and counterstained with 10 μL of DAPI-Antifade (DAPI in Fluorguard, 0.5 μg/mL; Insitus).
The slides were examined with an Olympus IX-50 microscope (Olympus, Tokyo, Japan) with these filters: blue filter cube (Arcturus, Mountain View, CA) for SpectrumGreen (centromeric probes for chromosomes 7, 17, and Y) Vysis probes, and the UV Filter (Chroma, Brattleboro, VT) for the DAPI nuclear counterstain. The signals were recorded with a CCD camera.
In Situ Hybridization Analysis
The criteria for evaluating the fluorescence in situ hybridization signals were adapted from Hopman et al. (19, 20) One hundred to 200 nuclei were scored for alpha-satellite signals observed with the fluorescence microscope at 400× magnification. As much as possible, signals from solitary nuclei were counted, but groups of two or three adjacent but not overlapping nuclei were occasionally included in the counts. Nuclei were counted when the entire nuclear circumference had a round-to-oval contour and showed no evidence of fragmentation. Two signals of the same size in close proximity, not connected by a link, were counted as two signals. A diffuse signal was regarded as a signal if it was contiguous and within an acceptable boundary. Two small signals connected by a visible link were counted as one signal. Overlapping nuclei and nuclei with uncertain signals were not counted. There was no significant variation in hybridization efficiency when different areas of the slides were examined.
RESULTS
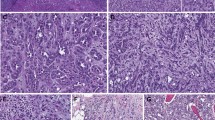
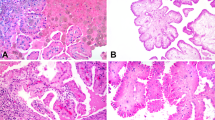
The tumors all had the typical histologic features of metanephric adenoma (Fig. 1). In normal epithelial cells in the renal tubules, nuclei with three or four signals were occasionally seen but in no sample exceeded 12% of the total. For chromosomes 7 and 17, 78% to 88% of the nuclei in each control sample showed two signals (median, 84%). Nuclei with only one signal for chromosome 7 or 17 invariably comprised <20% of the total nuclei counted. For the Y chromosome, 82 to 91% of the nuclei in each control sample showed a single signal.
In the seven metanephric adenomas, the results for chromosomes 7 and 17 (Table 1) were tightly clustered, and a high percentage (range, 75 to 85%; median, 79%) of nuclei showed two signals (Fig. 2). One metanephric adenoma had 19% of nuclei with three or more signals for chromosome 7. The Y chromosome was present in all three of the tumors from males (range, 86 to 89% of nuclei).
DISCUSSION
Brown et al. (9) studied 11 tumors by fluorescent in situ hybridization and reported gains of chromosomes 7 and 17 and loss of the Y chromosome in 8 tumors. The percentage of nuclei with three signals that they used as the threshold for recognizing trisomy was 7%, which some of the normal renal control tissues in the present study reached; they did not report counts on normal controls. Brown et al. (9) separately reported another case with a similar genetic pattern that was studied by classic cytogenetics (21). These findings are the chromosomal changes that have been reported most frequently in papillary renal cell neoplasia. Other reports of metanephric adenomas have included cytogenetic data from a small number of tumors and have generally shown normal karyotypes (2, 10, 11, 13, 14, 15, 22). In 1997, Renshaw et al. (22) described one tumor with a disomic pattern for chromosomes 7 and 17 and 3p and later reported a case of a child with a metastatic metanephric adenoma in which the karyotype was diploid, and a disomic pattern for chromosomes 7 and 17 was displayed by fluorescent in situ hybridization (14). Granter et al. (11) described normal karyotypes in two cases. Birgisson et al. (10) found a single tumor to be diploid on flow cytometry and to have a normal karyotype by fluorescent in situ hybridization analysis of chromosomes 7, 8 and 17. Tsuji et al. (15) reported one case with normal numbers of chromosomes 7 and 17.
These conflicting observations may be the result of methodologic differences; in particular, selecting a low threshold for recognizing chromosomal gains and not comparing the results with data from normal tissues studied at the same time could lead to overestimation of the frequency of chromosomal gains. Also, it is possible that Brown et al. (9) included some tumors that we would have considered papillary adenomas or papillary renal cell carcinomas. That article did not include any photomicrographs of any of the tumors with gains of chromosomes, so this remains speculation. In our study, we included only tumors that four of the authors (MB, JNE, GM, and LC) independently examined and agreed were typical metanephric adenomas, according to published criteria (8).
In this study, we observed in a series of seven tumors that metanephric adenoma lacks the frequent gains of chromosomes 7 and 17 and frequent losses of the Y chromosome that are typical of papillary renal cell neoplasms (23). The numbers of these chromosomes in the metanephric adenomas were similar to those that we observed in histologically normal epithelium of renal tubules. These findings suggest that metanephric adenoma is not related to papillary renal cell carcinoma and papillary adenoma. Analysis of chromosomes 7, 17, and Y by fluorescence in situ hybridization with centromeric probes could be useful in diagnostically differentiating metanephric adenoma with extensive papillary architecture and papillary renal cell carcinoma type 1.
References
Brisigotti M, Cozzutto C, Fabbretti G, Sergi C, Callea F . Metanephric adenoma. Histol Histopathol 1992; 7: 689–692.
Jones EC, Pins M, Dickersin GR, Young RH . Metanephric adenoma of the kidney, a clinicopathological, immunohistochemical, flow cytometric, cytogenetic, and electron microscopic study of seven cases. Am J Surg Pathol 1995; 19: 615–626.
Davis CJ Jr, Barton JH, Sesterhenn IA, Mostofi FK . Metanephric adenoma, clinicopathological study of fifty patients. Am J Surg Pathol 1995; 19: 1101–1114.
Drut R, Drut RM, Ortolani C . Metastatic metanephric adenoma with foci of papillary carcinoma in a child: a combined histologic, immunohistochemical, and FISH study. Int J Surg Pathol 2001; 9: 241–247.
Kovacs G, Akhtar M, Beckwith JB, et al. The Heidelberg classification of renal cell tumours. J Pathol 1997; 183: 131–133.
Störkel S, Eble JN, Adlakha K, et al. Classification of renal cell carcinoma, workgroup 1. Cancer 1997; 80: 987–989.
Pins MR, Jones EC, Martul EV, et al. Metanephric adenoma-like tumors of the kidney: report of 3 malignancies with emphasis on discriminating features. Arch Pathol Lab Med 1999; 123: 415–420.
Grignon DJ, Eble JN . Papillary and metanephric adenomas of the kidney. Semin Diagn Pathol 1998; 15: 41–53.
Brown JA, Anderl KL, Borell TJ, et al. Simultaneous chromosome 7 and 17 gain and sex chromosome loss provide evidence that renal metanephric adenoma is related to papillary renal cell carcinoma. J Urol 1997; 158: 370–374.
Birgisson H, Einarsson GV, Steinarsdottir M, Jonasson JG . Metanephric adenoma. Scand J Urol Nephrol 1999; 33: 340–343.
Granter SR, Fletcher JA, Renshaw AA . Cytologic and cytogenetic analysis of metanephric adenoma of the kidney, a report of two cases. Am J Clin Pathol 1997; 108: 544–549.
Renshaw AA, Zhang H, Corless CL, Fletcher JA, Pins MR . Solid variants of papillary (chromophil) renal cell carcinoma: clinicopathologic and genetic features. Am J Surg Pathol 1997; 21: 1203–1209.
Gatalica Z, Grujic S, Kovatich A, Petersen RO . Metanephric adenoma: histology, immunophenotype, cytogenetics, ultrastructure. Mod Pathol 1996; 9: 329–333.
Renshaw AA, Freyer DR, Hammers YA . Metastatic metanephric adenoma in a child. Am J Surg Pathol 2000; 24: 570–574.
Tsuji M, Murakami Y, Kanayama H-O, Sano T, Kagawa S . A case of renal metanephric adenoma: histologic, immunohistochemical and cytogenetic analyses. Int J Urol 1999; 6: 203–207.
Kattar MM, Grignon DJ, Wallis T, et al. Clinicopathologic and interphase cytogenetic analysis of papillary (chromophilic) renal cell carcinoma. Mod Pathol 1997; 10: 1143–1150.
Corless CL, Aburatani H, Fletcher JA, et al. Papillary renal cell carcinoma, quantitation of chromosomes 7 and 17 by FISH, analysis of chromosome 3p for LOH, and DNA ploidy. Diagn Mol Pathol 1996; 5: 53–64.
Kovacs G, Fuzesi L, Emanuel A, Kung H-F . Cytogenetics of papillary renal cell tumors. Genes Chromosomes Cancer 1991; 3: 249–255.
Hopman AHN, Ramaekers FCS, Raap AK, et al. In situ hybridization as a tool to study numerical chromosome aberrations in solid bladder tumors. Histochemistry 1988; 89: 307–316.
Hopman AH, Voorter CE, Ramaekers FC . Detection of genomic changes in cancer by in situ hybridization. Mol Biol Rep 1994; 19: 31–44.
Brown JA, Sebo TJ, Segura JW . Metaphase analysis of metanephric adenoma reveals chromosome Y loss with chromosome 7 and 17 gain. Urology 1996; 48: 473–475.
Renshaw AA, Maurici D, Fletcher JA . Cytologic and fluorescence in situ hybridization (FISH) examination of metanephric adenoma. Diagn Cytopathol 1997; 16: 107–111.
Brunelli M, Eble JN, Martignoni G, Zhang S, Cheng L . Gains of chromosomes 7, 17, 12, 16, and 20, and loss of Y occur early in the evolution of papillary renal cell neoplasia: a fluorescent in situ hybridization study. Mod Pathol 2003; 16.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by Fondazione Cassa di Risparmio di Verona (bando 2001); Diagnostica molecolare in oncologia (2003); Murst ex 60% (2003); and Banco di Sardegna.
Presented in part at the 92nd annual meeting of the U.S. and Canadian Academy of Pathology, March 25, 2003.
Rights and permissions
About this article
Cite this article
Brunelli, M., Eble, J., Zhang, S. et al. Metanephric Adenoma Lacks the Gains of Chromosomes 7 and 17 and Loss of Y That Are Typical of Papillary Renal Cell Carcinoma and Papillary Adenoma. Mod Pathol 16, 1060–1063 (2003). https://doi.org/10.1097/01.MP.0000090923.50509.55
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1097/01.MP.0000090923.50509.55
Keywords
This article is cited by
-
KANK1-NTRK3 fusions define a subset of BRAF mutation negative renal metanephric adenomas
BMC Medical Genetics (2020)
-
MYB-NFIB gene fusion in prostatic basal cell carcinoma: clinicopathologic correlates and comparison with basal cell adenoma and florid basal cell hyperplasia
Modern Pathology (2019)
-
Metanephric adenoma: the utility of immunohistochemical and cytogenetic analyses in differential diagnosis, including solid variant papillary renal cell carcinoma and epithelial-predominant nephroblastoma
Modern Pathology (2015)
-
Mucinous tubular and spindle cell carcinoma and solid variant papillary renal cell carcinoma: a clinicopathologic comparative analysis of four cases with similar molecular genetics datum
Diagnostic Pathology (2014)
-
Metanephric adenosarcoma: a rare case with immunohistochemistry and molecular analysis
Diagnostic Pathology (2014)