Abstract
We recently proposed that an epithelial renal tumor “papillary renal neoplasm with reverse polarity” represents a distinct entity. It constituted 4% of previously diagnosed papillary renal cell carcinoma at the participating institutions. Histologically, it is characterized by papillary or tubulopapillary architecture covered by a single layer of eosinophilic cells with finely granular cytoplasm and apically located nuclei. It is characteristically positive for GATA3 and L1CAM and lack vimentin and, to a lesser extent, α-methylacyl-CoA-racemase (AMACR/p504s) immunostaining. To investigate the molecular pathogenesis of these tumors, we performed targeted next-generation sequencing on ten previously reported papillary renal neoplasms with reverse polarity, followed by a targeted polymerase chain reaction analysis for KRAS mutations in a control series of 30 type 1 and 2 papillary renal cell carcinomas. KRAS missense mutations were identified in eight of ten papillary renal neoplasms with reverse polarity. These mutations were clustered in exon 2—codon 12: c.35 G > T (n = 6) or c.34 G > C (n = 2) resulting in p.Gly12Val and p.Gly12Arg alterations, respectively. One of the wild-type tumors had BRAF c.1798_1799delGTinsAG (p.Val600Arg) mutation. No KRAS mutations were identified in any of the 30 control tumors. In summary, this study supports our proposal that papillary renal neoplasm with reverse polarity is an entity distinct from papillary renal cell carcinoma and the only renal cell neoplasm to consistently harbor KRAS mutations.
Similar content being viewed by others
Introduction
In 1997, Delahunt and Eble proposed that papillary renal cell carcinoma could be classified into types 1 and 2 [1]. Results of the Cancer Genome Atlas (TCGA) molecular characterization have supported this histologic classification. However, the observed histologic heterogeneity of papillary renal cell carcinoma type 2 is also reflected on a molecular level as indicated by several studies [2,3,4,5]. Type 1 tumors are associated with MET alterations, whereas type 2 tumors consist of at least three subtypes based on molecular features and are characterized by CDKN2A silencing, SETD2 mutations, TFE3 fusions, and increased expression of the NRF2–antioxidant response element pathway [4]. Ultimately, these advances in genetic understanding have led to the recognition of distinct tumor entities that were once included under the umbrella of papillary renal cell carcinoma type 2, including for instance hereditary leiomyomatosis and renal cell carcinoma syndrome-associated renal cell carcinoma and some translocation carcinomas [6].
We recently described a subset of papillary renal neoplasms and proposed the term papillary renal neoplasm with reverse polarity [7]. This tumor was found equally in men and women, with a median age of 66 years. All 18 tumors were ≤3 cm in size (median 1.4 cm, mean 1.6 cm) and ten were encapsulated. All tumors were low stage and appear to have been cured by surgery. Microscopically, the tumors were composed of papillary or tubulopapillary architecture covered by a single layer of eosinophilic cells with finely granular eosinophilic cytoplasm and apically located round nuclei with inconspicuous nucleoli. No intracellular hemosiderin, psammoma bodies, mitotic figures, necrosis, or clusters of foamy macrophages were seen. The diagnosis was aided by positive immunohistochemical staining for GATA3 and L1CAM along with the lack of vimentin and, to a lesser extent, α-methylacyl-CoA-racemase (AMACR/p504s) staining. Fluorescence in situ hybridization analysis demonstrated a lower level of chromosomal abnormalities (chromosome 7 and 17 trisomy in 20% of tumors and deletion of Y chromosome in 14% of the tumors in males). These findings are largely atypical for papillary renal cell carcinomas types 1 and 2 [7].
In the current study, we performed mutational analysis of the papillary renal neoplasm with reverse polarity using a targeted next-generation sequencing (NGS) platform and report a novel recurrent genetic mutation in these tumors.
Materials and methods
Study population
The study was approved by the Institutional Review Board of the participating institutions (Indiana University, Indianapolis, IN; Henry Ford Health System, Detroit, MI; and Wayne State University, Detroit, MI). Based on availability of sufficient material for molecular analysis, the study included ten of the previously published tumors of papillary renal neoplasm with reverse polarity obtained from the three departments of pathology (corresponding tumors no. 2, 4, 5, 8, 10, 11, 12, 14, 15, and 17). As previously described, a control group composed of 30 papillary renal cell carcinomas (15 tumors of type 1 and 15 tumors of type 2) was selected from the archives of the participating institutions. To be selected for the control group, the tumors were reviewed independently by two pathologists (DJG and JNE). Both were blinded to the initial diagnosis and each other’s diagnosis. Only tumors where classification into types 1 and 2 was in agreement were then utilized as controls. All tumors had been fixed in 10% neutral-buffered formalin and embedded in paraffin [7].
NGS and mutational analysis
Representative formalin-fixed, paraffin-embedded blocks of ten papillary renal neoplasms with reverse polarity containing more than 60% tumor were submitted for targeted NGS using the University of Chicago Medicine OncoPlus (UCM-OncoPlus) panel, a hybrid-capture panel targeting 1213 cancer-associated genes. DNA extraction, DNA quantification, library preparation, and sequencing were performed as previously described [8]. Data analysis was performed on a compliant high-performance computing system (Center for Research Informatics, University of Chicago) using a University of Chicago developed bioinformatics pipeline. First, using Novoalign 3.02.07 (NovoCraft, Selangor, Malaysia), the data were aligned to the hg19 reference human genome. Then, Samtools 0.1.1915 mpileup and Variant Inspector version 1.0, an UCM-developed variant calling software, were used for variant calling. Subsequently, variants were filtered based on depth (>100×), Phred quality score (>30), minor allele frequency (MAF > 5%), and location in clinical exonic territory for review. Variant calls were annotated and converted to Human Genome Variation Society nomenclature using Alamut Batch version 1.3.0 software. Further manual variant curation was followed in accordance with the Association for Molecular Pathology recommendations [9].
KRAS mutational analysis
Based on the NGS findings, a targeted polymerase chain reaction analysis was performed on the control tumors as previously described [10]. Briefly, formalin-fixed, paraffin-embedded tissue blocks were retrieved from each control tumor. DNA extraction from each block was performed using the Qiagen QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA, USA). DNA concentration was determined using the NanoDrop Spectrophotometer and adjusted to ~10 ng/μl in ddH2O. Polymerase chain reaction testing was performed according to the recommended procedure using the Qiagen therascreen KRAS RGQ PCR Kit on the Qiagen Rotor-Gene Q MDx instrument. The therascreen KRAS RGQ PCR Kit provides eight separate PCR amplification reactions: seven mutation-specific reactions in codons 12 and 13 of exon 2 of the KRAS oncogene [Gly12Ala (G12A), Gly12Asp (G12D), Gly12Arg (G12R), Gly12Cys (G12C), Gly12Ser (G12S), Gly12Val (G12V), and Gly13Asp (G13D)] and a wild-type control in exon 4. Analysis of crossing thresholds and mutation calls for each PCR amplification reaction were performed by the Rotor-Gene Q therascreen KRAS Assay analysis software once runs were completed.
Evaluation of TCGA renal neoplasms data for gene expression
Data of the mRNA expression (mRNA seqV2, RSEM normalized values) of a cohort of 17 tumors obtained from TCGA(Kidney renal papillary cell carcinoma “KIRP” project) were utilized for mRNA clustering analysis using 20531 gene signatures. The tumors were divided into four groups based on the KRAS mutational status and the histologic criteria, as defined by Delahunt and Eble [1], using the digital images available at TCGA website. These groups included: three papillary renal neoplasms with reverse polarity (TCGA-2Z-A9JN, TCGA-BQ-5883, and TCGA-A4-8312), two KRAS mutated papillary renal cell carcinomas (TCGA-GL-A9DC and TCGA-MH-A854), six papillary renal cell carcinomas type 1 (TCGA-2Z-A9J6, TCGA-2Z-A9JE, TCGA-2Z-A9JL, TCGA-2Z-A9JZ, TCGA-2Z-A9K0, and TCGA-SX-A71S), and six papillary renal cell carcinomas type 2 (TCGA-2Z-A9J2, TCGA-2Z-A9JD, TCGA-5P-A9JW, TCGA-B9-A8YI, TCGA-B9-A44B, and TCGA-IA-A83S).
Unsupervised clustering analysis of the 17 tumors was performed using the GENE-E software (https://software.broadinstitute.org/GENE-E/index.html/) and Genesis platform (Graz, Austria) [11]. K-mean clustering with average linkage of Euclidian distance for both samples and genes was performed. Same set of samples was analyzed in Firebrowse portal (http://www.firebrowse.org/).
Results
NGS was performed on DNA isolated from ten papillary renal neoplasms with reverse polarity as described in the section “Materials and methods.” The clinicopathologic and mutational results are summarized in Table 1 and Supplementary Table 1. Bioinformatics analysis revealed recurrent missense mutations in the KRAS (NM_033360.3) in eight tumors (80%). These mutations were due to either a c.35 G > T (n = 6) or c.34 G > C (n = 2) substitution, resulting in p.Gly12Val and p.Gly12Arg alterations, respectively (Fig. 1). The minor allele frequency ranged from 13–34%, supporting the somatic origin. Both of these variants are common somatic mutations in cancer and have been classified as pathogenic in the Catalogue of Somatic Mutations in Cancer (COSMIC) and ClinVar databases [12, 13].
Integrative Genomics Viewer snapshot of human KRAS gene with the location of the missense mutations identified in eight papillary renal neoplasms with reverse polarity tumors. The mutations are all clustered within two recurrent hotspots (p.Gly12Val and p.Gly12Arg). Blue denotes T-nucleotide (normal), green denotes A-nucleotide (p.Gly12Val), and orange denotes G-nucleotide (p.Gly12Arg)
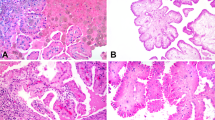
Targeted NGS also revealed mutations in TP53, BRCA2, and BRAF in tumors no. 3, 8, and 9, respectively, and duplication in BRCA2 in tumor no. 1 (Fig. 2). BRCA2 is a tumor suppressor gene and is known to predispose patients to breast and ovarian carcinoma [14, 15]. A frameshift duplication c.5073dup (p.Trp1692Metfs*3) and missense mutations c.4258 G > T (p.Asp1420Tyr) were identified in the BRCA2 gene (NM_000059.3) in two tumors (no. 1 and 8, respectively). Both have been reported as pathogenic somatic mutations in cancer (COSMIC v89) [16, 17]. Tumor no. 3 harbored missense mutation in the TP53 (NM_000546.5) tumor suppressor gene c.638 G > A (p.Arg213Gln). This mutation is recurrent in many sporadic and familial tumors [18]. Lastly, one tumor (no. 9) carried BRAF (NM_004333) c.1798_1799delGTinsAG (p.Val600Arg) mutation, which is reported in melanoma, colorectal, and pancreatic carcinoma (COSM474) [16, 17]. This tumor lacked KRAS mutation. BRAF protein is a downstream effector in the KRAS pathway and the mutual exclusivity of these two mutations has been suggested by some researchers [19, 20]. Representative histologic images of the analyzed tumors are shown in Fig. 3.
Representative histologic images of papillary renal neoplasms with reverse polarity. All formed thin papillary architecture with delicate fibrovascular cores with variable proportion of tubular formation. The cells are mostly cuboidal with eosinophilic, finely granular cytoplasm and the nuclei are characteristically located at the apical surface of the cells. Nuclei are mostly small, nonoverlapping with regular nuclear contours, occasional nuclear clearing, and inconspicuous nucleoli. a, b KRAS and BRCA1 mutated (tumor no. 8) or BRCA1 duplicated (tumor no. 1). c KRAS only mutated (tumor no. 2). d KRAS and TP53 mutated (tumor no. 3). e BRAF mutated (tumor no. 9). f KRAS wild type (tumor no. 10)
We performed a targeted polymerase chain reaction analysis of the 30 papillary renal cell carcinomas in the control group and found no hotspot mutations in the KRAS in any of them.
To further validate our findings, we examined the publicly available database in TCGA (KIRP project) and found five tumors with KRAS mutations. Three of the five were papillary renal neoplasms with reverse polarity (TCGA-2Z-A9JN, TCGA-A4–8312, and TCGA-BQ-5883) based on the available digital images (Figs. 4 and 5). All of these had KRAS mutations in the same hotspots (c.35 G > T and c.34 G > C) found in our tumors. Interestingly, the mutational burden of the three tumors was low (≤20 mutations), with KRAS being the only shared mutation among them. All three tumors were reported as diploid.
a, b, c The Cancer Genome Atlas histologic images of the three papillary renal neoplasms with reverse polarity tumors harboring KRAS mutation [(TCGA-2Z-A9JN, TCGA-BQ-5883, and TCGA-A4-8312, respectively), https://portal.gdc.cancer.gov/projects/TCGA-KIRP]
a, b The Cancer Genome Atlas histologic images of the two KRAS mutated papillary renal cell carcinomas, type 1 [(TCGA-GL-A9DC and TCGA-MH-A854, respectively), https://portal.gdc.cancer.gov/projects/TCGA-KIRP]
Unsupervised mRNA clustering of the 17 Cancer Genome Atlas tumors (three papillary renal neoplasms with reverse polarity, two KRAS mutated papillary renal cell carcinomas, six papillary renal cell carcinomas type 1, and six papillary renal cell carcinomas type 2) showed papillary renal neoplasms with reverse polarity to have a different expression profile compared with other groups (Fig. 6), further supporting our analysis.
Unsupervised mRNA clustering analysis of 17 tumors obtained from the Cancer Genome Atlas database [three papillary renal neoplasms with reverse polarity (PRNRP), two KRAS mutated papillary renal cell carcinomas (PRCC-KRAS-MUT), six papillary renal cell carcinomas type 1 (PRCC-1), and six papillary renal cell carcinomas type 2 (PRCC-2)] using 20531 genes. Papillary renal neoplasm with reverse polarity tumors cluster together in a distinct group. The clusters shown are based on K-mean method (GENE-E) with average linkage of Euclidian distance for both samples and genes. Blue denotes upregulated genes and red denotes downregulated genes
Discussion
This study identifies for the first time a renal cell neoplasm with consistent KRAS mutations. In addition to our previous report of the distinct morphologic, immunohistochemical, and chromosomal features of papillary renal neoplasm with reverse polarity, this study also shows a recurrent pathogenic mutation that is distinct from any previously found in renal cell neoplasms, including papillary renal cell carcinoma types 1 and 2.
KRAS proto-oncogene, GTPase is part of the RAS family. It encodes GTPase enzyme that functions as molecular regulator of proliferation and cell survival pathways [21]. Abnormal function of KRAS is associated with tumorigenesis and typically arises from single mutation at codons 12, 13, or 61, which leads to constitutive activation of the gene [22].
In addition to papillary renal neoplasm with reverse polarity, the COSMIC v89 shows KRAS mutation in a number of cancers, including pancreatic, colorectal, and lung adenocarcinomas. KRAS mutations are also reported in benign lesions, although to a much lesser extent than cancers, including papillary fibroelastoma, ovarian mucinous cystadenoma, and intracranial arteriovenous malformations [10, 23, 24]. In the kidney, it accounts for only a minority (0.8%, n = 33/4207) of mutations reported in COSMIC (Supplementary Table 2). Five (5/33) of these tumors were reported as papillary renal cell carcinomas, two of which are papillary renal neoplasms with reverse polarity as determined by examining the available digital histologic images (TCGA-BQ-5883-01 and TCGA-A4-8312-01). Of note, one of the studies that were cited in COSMIC v89 reported KRAS mutations in 100% of clear cell renal cell carcinomas (n = 11) [25]. These results have not been reproduced by larger studies including TCGA database where only 0.1% of clear cell renal cell carcinomas had a KRAS mutation, n = 1/537 in the kidney renal clear cell carcinoma project.
In a report by Saleeb et al., a partial molecular overlap between tumors similar to papillary renal neoplasm with reverse polarity and papillary renal cell carcinoma type 2 was found, where papillary renal cell carcinoma type 2 had additional unique pathways associated with tumor aggressiveness and metastasis. However, when clustering analysis was performed, papillary renal neoplasm with reverse polarity tumors had its own distinct molecular cluster with minimal overlap with papillary renal cell carcinoma type 2 [3]. Further, similar clusters were found based on KRAS mutational status of the same Cancer Genome Atlas tumors in Firebrowse portal (Fig. 7).
KRAS mutation status versus mRNA Seq hierarchical from Firebrowse portal. The three papillary renal neoplasms with reverse polarity tumors are clustered into distinct cluster than other KRAS mutated tumors. Broad Institute TCGA Genome Data Analysis Center (2016): correlation between gene mutation status and molecular subtypes. Broad Institute of MIT and Harvard. doi:10.7908/C1PN9526
The shared characteristic location of the nuclei away from the basement membrane is a unique feature, which has been described in only a few tumors, including tall cell carcinoma (solid papillary carcinoma) with reverse polarity of the breast. Chiang et al. were the first to describe a unique IDH2 or PIK3CA mutation in these tumors [26]. However, mutations in those two genes were not identified in our tumors, suggesting that another driving genes responsible for the reverse polarity phenotype.
In summary, the identification of consistent pathogenic KRAS mutations in papillary renal neoplasm with reverse polarity provides further evidence of the distinct nature of this entity. Although further studies with a larger series of tumors are recommended, the finding of KRAS mutation is two separate, unrelated cohorts (ours and TCGA), which supports our proposal that this morphologic entity is a result of a molecular pathogenic process different from other renal cell neoplasms. Our study provides the evidence for the first renal cell neoplasm characterized by KRAS mutation.
References
Delahunt B, Eble JN. Papillary renal cell carcinoma: a clinicopathologic and immunohistochemical study of 105 tumors. Mod Pathol. 1997;10:537–44.
Marsaud A, Dadone B, Ambrosetti D, Baudoin C, Chamorey E, Rouleau E, et al. Dismantling papillary renal cell carcinoma classification: the heterogeneity of genetic profiles suggests several independent diseases. Genes Chromosomes Cancer. 2015;54:369–82.
Saleeb RM, Brimo F, Farag M, Rompre-Brodeur A, Rotondo F, Beharry V, et al. Toward biological subtyping of papillary renal cell carcinoma with clinical implications through histologic, immunohistochemical, and molecular analysis. Am J Surg Pathol. 2017;41:1618–29.
Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Cancer Genome Atlas Research Network, et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med.2016;374:135–45.
Saleeb RM, Plant P, Tawedrous E, Krizova A, Brimo F, Evans AJ, et al. Integrated phenotypic/genotypic analysis of papillary renal cell carcinoma subtypes: identification of prognostic markers, cancer-related pathways, and implications for therapy. Eur Urol Focus. 2018;4:740–8.
Srigley JR, Delahunt B, Eble JN, Egevad L, Epstein JI, Grignon D, et al. The International Society of Urological Pathology (ISUP) Vancouver classification of renal neoplasia. Am J Surg Pathol. 2013;37:1469–89.
Al-Obaidy KI, Eble JN, Cheng L, Williamson SR, Sakr WA, Gupta N, et al. Papillary renal neoplasm with reverse polarity: a morphologic, immunohistochemical, and molecular study. Am J Surg Pathol. 2019;43:1099–111.
Parilla M, Alikhan M, Al-Kawaaz M, Patil S, Kadri S, Ritterhouse LL, et al. Genetic underpinnings of renal cell carcinoma with leiomyomatous stroma. Am J Surg Pathol. 2019;43:1135–44.
Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23.
Priemer DS, Vortmeyer AO, Zhang S, Chang HY, Curless KL, Cheng L. Activating KRAS mutations in arteriovenous malformations of the brain: frequency and clinicopathologic correlation. Hum Pathol. 2019;19:33–9.
Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–8.
Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–83.
Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–67.
Antoniou AC, Pharoah PD, McMullan G, Day NE, Ponder BA, Easton D. Evidence for further breast cancer susceptibility genes in addition to BRCA1 and BRCA2 in a population-based study. Genet Epidemiol. 2001;21:1–18.
Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Kwan E, et al. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet. 2001;68:700–10.
Dutton-Regester K, Kakavand H, Aoude LG, Stark MS, Gartside MG, Johansson P, et al. Melanomas of unknown primary have a mutation profile consistent with cutaneous sun-exposed melanoma. Pigment Cell Melanoma Res. 2013;26:852–60.
Zhu XL, Cai X, Zhang L, Yang F, Sheng WQ, Lu YM, et al. [KRAS and BRAF gene mutations in correlation with clinicopathologic features of colorectal carcinoma in Chinese]. Zhonghua Bing Li Xue Za Zhi 2012;41:584–9.
Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–63.
Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54.
Oikonomou E, Koustas E, Goulielmaki M, Pintzas A. BRAF vs RAS oncogenes: are mutations of the same pathway equal? Differential signalling and therapeutic implications. Oncotarget. 2014;5:11752–77.
Quinlan MP, Settleman J. Isoform-specific ras functions in development and cancer. Future Oncol. 2009;5:105–16.
Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–67.
Wittersheim M, Heydt C, Hoffmann F, Buttner R. KRAS mutation in papillary fibroelastoma: a true cardiac neoplasm? J Pathol Clin Res. 2017;3:100–4.
Lee YJ, Lee MY, Ruan A, Chen CK, Liu HP, Wang CJ, et al. Multipoint Kras oncogene mutations potentially indicate mucinous carcinoma on the entire spectrum of mucinous ovarian neoplasms. Oncotarget. 2016;7:82097–103.
Raspollini MR, Castiglione F, Martignoni G, Cheng L, Montironi R, Lopez-Beltran A. Unlike in clear cell renal cell carcinoma, KRAS is not mutated in multilocular cystic clear cell renal cell neoplasm of low potential. Virchows Arch. 2015;467:687–93.
Chiang S, Weigelt B, Wen HC, Pareja F, Raghavendra A, Martelotto LG, et al. IDH2 mutations define a unique subtype of breast cancer with altered nuclear polarity. Cancer Res. 2016;76:7118–29.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Al-Obaidy, K.I., Eble, J.N., Nassiri, M. et al. Recurrent KRAS mutations in papillary renal neoplasm with reverse polarity. Mod Pathol 33, 1157–1164 (2020). https://doi.org/10.1038/s41379-019-0362-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-019-0362-1
This article is cited by
-
Ipsilateral synchronous papillary renal neoplasm with reverse polarity and urothelial carcinoma in a renal transplant recipient: a rare case report with molecular analysis and literature review
Diagnostic Pathology (2023)
-
Genomic alterations and diagnosis of renal cancer
Virchows Archiv (2023)
-
Papillary renal neoplasm with reverse polarity may be a novel renal cell tumor entity with low malignant potential
Diagnostic Pathology (2022)
-
Papillary renal cell carcinoma: a single institutional study of 199 cases addressing classification, clinicopathologic and molecular features, and treatment outcome
Modern Pathology (2022)
-
Recurrent KRAS mutations are early events in the development of papillary renal neoplasm with reverse polarity
Modern Pathology (2022)