Abstract
Although recent studies have suggested that p16INK4a may be a useful surrogate biomarker of cervical neoplasia, Ki-67 and human papillomavirus testing have also been shown to be useful in detecting neoplasia. To help delineate the utility of p16INK4a, biopsy samples (n = 569: negative, 133; reactive, 75; atypical, 39; low grade, 76; moderate, 80; and severe intraepithelial neoplasia, 113; also, squamous cell carcinoma, 46; adenocarcinoma, 7) were analyzed by immunohistochemistry for expression of p16INK4a and Ki-67 (n = 432), as well as by in situ hybridization for human papillomavirus Type 16 (n = 219). Testing for high-risk human papillomavirus types by polymerase chain reaction and HybridCapture2 was performed on concurrent cervical swab specimens. Recuts of the original blocks were reexamined (n = 198). Endometrial biopsies (n = 10) were also analyzed for p16INK4a expression. Degree of p16INK4a and Ki-67 expression correlated with degree of cervical neoplasia (P < .001) and with presence of high-risk human papillomavirus types (P < .001). There was no relationship between p16INK4a overexpression and inflammation or hormonal status. Ki-67 expression correlated with inflammation (P = 0.003) and was expressed in more reactive and atypical lesions than p16INK4a (P = 0.008). Probes for human papillomavirus 16 stained 54% of cervical neoplastic lesions; the degree of staining correlated significantly with degree of neoplasia (P < .001) and p16INK4a staining (P < .001). Interobserver reproducibility was substantial for p16INK4a and Ki-67 interpretation (weighted κ: 0.74 and 0.70, respectively). Expression of p16INK4a was observed in all endometrial biopsies. Compared with Ki-67 expression and detection of high-risk human papillomavirus, p16INK4a was less likely to be positive in samples from women with negative, reactive, and atypical biopsies. Although expression of p16INK4a in endometrial epithelium may be problematic in terms of screening, the potential of p16INK4a as a screening test warrants investigation.
Similar content being viewed by others
INTRODUCTION
The screening of women by Pap smear has led to a remarkable decline in the mortality from cervical cancer; however, secondary to subjective criteria, interpretation of Pap smears is subject to marked inter- and intraobserver variability as well as having a relatively low sensitivity for cervical neoplasia on a single sample (as low as 66% sensitivity for biopsy-proven high-grade squamous intraepithelial lesions [HSIL]) (1, 2). Recently, histology, which is thought of as the gold standard for the diagnosis of cervical neoplasia, has also been found to suffer from marked intra- and interobserver variability, and testing for high-risk human papillomavirus (HPV) by Hybrid Capture 2, which has been shown to be very sensitive in the detection of cervical neoplasia and useful in the triaging of ASCUS smears, has a low specificity for cervical neoplasia (1, 3). Thus, new biomarkers that are more sensitive and specific in the detection of cervical neoplasia and more reproducible than cervical cytology are needed.
Human papillomaviruses (HPV) are known to be a major causative agent in cervical neoplasia and invasive cervical carcinoma. Many different HPV types associated with cervical neoplasia have been discovered, and they have been subdivided into high- and low-risk categories based on their association with invasive cervical carcinoma (4). This association is based, in part, on the relative affinity that the HPV-type specific oncoproteins E6 and E7 bind to cellular regulatory proteins, specifically, the p53 tumor suppressor protein and the retinoblastoma protein (Rb) (5). Inactivation of these factors, either by degradation (p53) or functional inactivation (Rb), leads to disruption of the cell cycle and increased proliferation, thought to ultimately give rise to carcinoma.
p16INK4a is a cyclin-dependent kinase inhibitor that regulates the activity of cyclin-dependent kinases 4 and 6 and is often inactivated in many cancers by genetic deletion or hypermethylation (6). In non-HPV–associated tumors, this inactivation leads to increased cyclin-dependent kinase activity and inactivation of Rb. However, in HPV-associated tumors, inactivation of Rb by E7 leads to markedly increased levels of p16INK4a. Recent studies have documented overexpression of p16INK4a not only in cervical intraepithelial neoplasia (CIN) but in cervical cancer as well (6, 7, 8, 9, 10).
The purpose of this study was to (1) compare the relationship between the degree of neoplasia present and the intensity of p16INK4a staining, (2) analyze expression of p16INK4a in cervical biopsy specimens covering the diagnostic spectrum from negative to invasive carcinoma, (3) evaluate p16INK4a in comparison to Ki-67 immunohistochemistry, and (4) correlate p16INK4a staining with the presence of high-risk HPV types (determined by HPV16 in situ hybridization on tissue samples and polymerase chain reaction [PCR] or HybridCapture2 in concurrent cervical swab specimens). Additionally, as p16INK4a has potential as a screening test, endometrial tissue was analyzed for p16INK4a expression because endometrial cells are often found in cervicovaginal cytology samples.
MATERIALS AND METHODS
Case Selection
Cervical biopsy/LEEP (loop electrical excision procedure) specimens (n = 597) were obtained from an ongoing study at the University of Washington, the details of which are specified elsewhere (11). In brief, between December 1997 and October 2000, 4358 women from three Planned Parenthood clinics in Washington state were eligible to participate. Patients enrolled in the study received a gynecological exam, were provided cervical cytology, and were tested for high-risk HPV by Hybrid Capture2 and PCR-based assay. Women with an atypical squamous cells of undetermined significance (ASCUS), low-grade squamous intraepithelial lesion (LSIL), or ≥high grade SIL (HSIL) on liquid-based Pap test, or with a positive PCR or Hybrid Capture2 test for high risk HPV types (even with a negative Pap smear), were referred for colposcopy and biopsy. A random sample of women with negative HPV deoxyribonucleic acid (DNA) and cytology results was also referred to colposcopy and biopsy. Repeat HPV testing was performed by both Hybrid Capture2 on Pap test specimens and by PCR-based testing of cervical swab samples that had been placed in standard transport media when taken at the colposcopy visit (11). The 597 specimens were obtained from 382 different women, of whom 236 contributed only one sample and 146 contributed more than one sample (88 contributed two samples, 47 contributed three samples, and 11 contributed four samples).
Additionally, biopsy samples of invasive cervical carcinoma (n = 48) were obtained from another ongoing study on cervical cancer, this one among African women and conducted in cooperation with the University of Dakar in Senegal. PCR testing for HPV was performed on cervical swab specimens collected in standard transport media at the time of biopsy. The details of the study are specified elsewhere (12).
The endometrial biopsy samples were randomly selected from the Division of Pathology database at Harborview Medical Center.
All biopsy specimens were fixed with 10% neutral buffered formalin, and sections were processed by conventional methods. Original diagnoses were obtained on the initial H&E slides. In the tables and for analysis, the atypical category encompassed lesions diagnosed as follows: atypical, not otherwise specified (NOS); atypical, favor reactive; and atypical squamous metaplasia. (These are analyzed separately according to the different diagnoses in the Results section.) After additional levels had been obtained for immunohistochemistry, an extra section was cut and stained with H&E (“recut,” n = 198) and analyzed by one of the authors (SNA) for the following: (1) the presence of neoplasia, (2) degree of inflammation, (3) the location of the lesion, and (4) the presence of immature squamous metaplasia.
Immunohistochemistry
Immunohistochemistry for p16INK4a and Ki-67 was performed on 569 and 432 samples, respectively. Four-micrometer sections of formalin fixed paraffin embedded tissue were cut and placed on Superfrost Plus microscope slides. The tissue sections were then deparaffinized and rehydrated through graded alcohols. Endogenous peroxidase activity was blocked by an incubation in 3% H2O2. Antigen retrieval was carried out with 0.01 m citrate buffer pH 6.0 and microwave heat induction (13). MTM Laboratories (Heidelberg, Germany) supplied the p16INK4a antibody. The antibody is directed against the human p16INK4a tumor suppressor protein. The clone designation is E6H4 and identifies an epitope that is between aa134–156 of p16INK4a. The monoclonal antibody clone MIB-1 from DAKO Corporation (Carpenteria, CA) was used to detect the Ki-67 antigen. The native Ki-67 antigen and the recombinant fragments of the molecule are both detected by this antibody. The dilution of the p16INK4a and Ki-67 antibodies was 1:800 and 1:50, respectively. Approximately 100 microliters of the primary antibody was applied to each slide. The slides were washed and a biotinylated anti-mouse antibody was applied. After another wash, the avidin-biotin-peroxidase complex was applied. Color development was accomplished by incubation in diaminobenzidine with 3% H2O2 as a substrate. Nickel chloride was used to enhance and modify the color of the diaminobenzidine reaction product. The slides were counterstained in methyl green, dehydrated through graded alcohols, cleared in xylene, and coverslipped with permanent mounting media. Internal or external positive controls (for p16INK4a, CIN III lesions; for Ki-67, inflammatory cells) and negative controls (substitution of mouse ascites fluid for the primary antibody) were included with every run
For p16INK4a, the results were reported in semiquantitative fashion (negative, or 1+ to 3+) based on none, 5–25%, 25–75%, and >75% of cells immunostained in a lesion. Strong nuclear as well as cytoplasmic staining was considered a positive reaction. Wispy weak cytoplasmic staining present in rare cells (<5%) was considered ±, and for analysis was grouped into the negative category. For Ki-67, the results were also reported in a semiquantitative fashion as cells in the lower 1/3 of the epithelium staining (i.e., usually basilar cell staining), cells in the middle 1/3–2/3 staining, or cells in the upper 1/3 staining (14). Strong nuclear staining was considered a positive reaction. Stains were analyzed by two authors (SNA and JM) for reproducibility; each was blinded to the other’s result.
In Situ Hybridization
Four-micrometer sections were cut and placed onto Probe On Plus slides. The slides were baked at 60° C, then deparaffinized in xylene and rehydrated through a series of graded alcohols. Endogenous peroxidase activity was blocked by incubation in 3% hydrogen peroxide. The slides were heated in a microwave oven in 10 mm citrate buffer (15). Tissue digestion was carried out with pepsin. The slides were postfixed in paraformaldehyde and dehydrated through a series of graded alcohols. A biotinylated HPV 16 DNA probe (DAKO) was used. The HPV 16 probes target regions of the E1, E2, E4, E5, E6, and E7 open reading frames and the upstream regulatory region. The probe was applied to the tissue section, and hybridization was carried out at 37° C overnight. Unbound and mismatched hybrids were removed in a series of stringency washes. Visualization was via horseradish peroxidase (HRP) conjugated streptavidin with tyramide signal amplification, diaminobenzidine (3,3′-diaminobenzidine) as the chromogen, and hydrogen peroxide as the substrate (with nickel chloride enhancement) (16). The slides were counterstained in methyl green and examined by light microscopy.
HPV in situ hybridization was graded in a semiquantitative fashion (negative or 1+ to 3+) based on the percentage of cells staining within a lesion (none, <25%, 25–75%, or >75%). Positive controls (known HPV 16–positive lesions by PCR) and negative controls (hybridization mixture without the labeled probe) were included with each run.
HPV DNA Testing
Before PCR assay, DNA isolation and purification was carried out using the QIAamp DNA Mini Kit (QIAGEN Ltd, Crawley, UK) per the manufacturer’s instructions. HPV DNA PCR amplification reactions were then performed using 5′ biotinylated MY09, MY11, and HMB01 primers and Amplitaq Gold polymerase. To prevent PCR product carryover, dTTP was replaced by dUTP and Uracil-N-glycosylase (UNG) was added. The human β-globin gene was coamplified in the HPV reaction mix using 5′ biotinylated primers PC04 and GH20 to monitor specimen adequacy. Two μL of each specimen was added to 100 μL of reaction mix. PCR amplification was carried out in a Perkin Elmer Thermal Cycler 9600 with the following profile: 95° C for 9 minutes to activate the Amplitaq Gold; 40 cycles of 95° C for 1 minute, 55° C for 1 minute, and 72° C for 1 minute each, and a 5-minute terminal extension at 72° C.
HPV DNA typing analysis was performed (according to the manufacturer’s specifications) using the reverse-line strip test (Roche, Emeryville, CA) to detect high-risk HPV 16,18,26,31,33,35,39,45,51,52,55,56,58,59,68,73,82, 84 (17). Appropriate positive and negative controls were included with each run.
The Hybrid Capture 2™ (HC2) test (Digene Corp, Gaithersburg, MD), which was configured to detect in a single assay one or more of the following high-risk HPV types: HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, was performed (according to the manufacturer’s specifications) on samples taken concurrently with the biopsy (18).
Statistical Analysis
χ2 tests were used to assess associations between variables. The Kappa statistic (weighted and unweighted) was used to assess degree of interobserver agreement. Association of variables of interest (p16INK4a, Ki-67, etc.) were considered significant if the two-sided χ2 test had a P value of <.05. Analyses stratified by HPV status or other potentially confounding variables were assessed using Mantel-Haenszel analysis. Statistical analysis was conducted using SAS (Cary, NC).
A number of individuals contributed more than one sample to the study analysis, either by providing samples during different visits, or multiple slice/sections of biopsy tissue from the same visit, or both. To account for potential bias caused by women who contributed more than one sample, we performed a regression analysis to adjust for correlated data using Generalized Estimating Equations SAS procedure GENMOD. The adjusted analysis did not differ from the unadjusted analysis, so we present results from the unadjusted analysis.
RESULTS
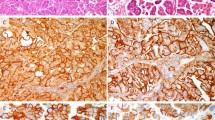
The degree of p16INK4a expression correlated well with the degree of cervical neoplasia, and this correlation improved slightly when compared with the recut slide diagnosis (P < .001; Fig. 1; Tables 1 and 2). There was very little expression in negative and reactive lesions, with only 11% to 12% showing ≥1+ staining (24 of 208 -original diagnosis, 12 of 112 recut diagnosis). 57% of the CIN I cases had ≥1+ expression, compared with 75% of CIN II lesions and 91% of CIN III lesions. On the recut diagnosis, 97% of CIN III lesions stained ≥1+. There were 10 (9%) CIN III original diagnosis that did not stain for p16INK4a, but on review, the majority of these were secondary to the lesion being cut through and not present on the immunohistochemistry (IHC) slide. For the recut diagnosis, there was only 1 (3%) case that did not stain with p16INK4a, and on review, two of three pathologists agreed that this represented CIN III, whereas the third felt it represented atypical squamous metaplasia. p16INK4a expression of 1+ or greater was present in 89%(47/53) of the invasive carcinomas. Review of negative cases confirmed the carcinoma diagnosis.
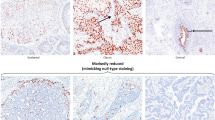
Fifty-three of the 571 cases had ± staining (as defined above, wispy faint cytoplasmic staining in rare cells [<5%]). This staining did not resemble the true positive staining and on analysis was found to significantly correlate with the presence of inflammation (P = .006). Because ± p16INK4a staining did not resemble true positive nuclear and cytoplasmic staining, was not associated with the detection of high-risk HPV, and was strongly related to detection of inflammation, it was considered a negative finding for p16INK4a staining.
Of the atypical lesions (n = 39), 6 (15%) of the original diagnosis showed ≥1+ p16INK4a expression compared with 4 (31%) of the recut diagnosis. Two of these were originally coded as atypical squamous metaplasia (ASM), one atypical favor low-grade dysplasia, one atypical not otherwise specified, and two atypical favor reactive. Of the atypical lesions with p16INK4a expression, most (4 of 6) had either a history of significant dysplasia, concurrent dysplasia, were high-risk HPV positive, and/or had dysplasia on follow-up. Ki-67 was elevated (intermediate expression) in 2 of 4 of the cases. One case of atypical squamous metaplasia that was p16INK4a positive had a previous biopsy and Pap that were diagnosed as high-grade dysplasia but only showed atypical squamous metaplasia on the LEEP specimen. The other case of p16INK4a positive atypical squamous metaplasia lacked high-risk HPV, and the concurrent Pap was negative; unfortunately no follow-up was available for this case.
Ki-67 expression also significantly correlated with the degree of cervical neoplasia (P < .001; Fig. 1; Table 3). The expression pattern was similar to that for p16INK4a in that in CIN I, expression was usually confined to the middle one third of the epithelium, and in CIN III, expression was often present in the upper one third. However, one of the major differences was that Ki-67 expression in the middle one third of the epithelium or higher was present in 29% of both reactive and atypical lesions, whereas only 13% (10/75) of reactive and 15% (6/39) of atypical lesions had ≥1+ expression of p16INK4a. This difference was statistically significant (P = .008). The intermediate or greater expression of Ki-67 also correlated significantly with inflammation (P = .003), whereas ≥1+ expression of p16INK4a did not correlate with inflammation. The portion of originally diagnosed CIN I lesions expressing Ki-67 above the lower one third of the epithelium was 60%; however, the portion of recut diagnosed CIN I lesions expressing Ki-67 above the lower one third was 83% (10 of 12), implying a significant degree of lesion cut-through in the CIN I lesions. Ki-67 expression was present in 92% (47/51) invasive carcinomas.
HPV 16 in situ hybridization was positive in 54% of the neoplastic lesions overall, and the degree of hybridization also correlated significantly with the degree of neoplasia (P < .001) and p16INK4a staining (P < .001; Table 4). Of negative and reactive lesions, 9% and 14%, respectively, were positive for HPV 16 in situ hybridization. The majority of samples (68%) that showed 3+ p16INK4a expression stained positively for HPV 16, and only 12 of 54 (22%) biopsies positive for HPV 16 by in situ hybridization lacked p16INK4a expression. Only 50% of low-grade lesions (12/24) were positive for HPV 16, as opposed to 64% (32/50) of CIN III lesions.
p16INK4a staining correlated significantly with the presence of high-risk HPV types as measured by HybridCapture2 or PCR-based testing of cervical swab samples taken concurrently at the biopsy/colposcopy visit (Table 5). The majority of CIN lesions positive for p16INK4a were also positive for high-risk HPV by HybridCapture2 (84%); however, HybridCapture2 was positive in 17 (36%) histologically negative cases that were negative with p16INK4a. PCR results were similar to HybridCapture2 results in that 90% of CIN III cases were positive for both p16INK4a and high-risk HPV, but PCR was also positive for high-risk HPV in 47% of histologically negative cases.
In the invasive carcinoma samples, p16INK4a expression was more often detected than high-risk HPV DNA (Table 6). p16INK4a expression was present in 89% of the invasive cervical carcinomas, whereas PCR-based assay only detected HPV DNA in 73% of the cases.
There was no significant difference between p16INK4a expression in samples positive for HPV 16 versus other high-risk HPV types and no difference in expression in samples positive for one versus multiple high-risk HPV types.
In the endometrial samples, p16INK4a was strongly expressed in the epithelium of all the samples tested: secretory (4/4), proliferative (3/3), and hyperplastic (1/1) endometrium, as well as endometrial adenocarcinoma (2/2; Fig. 2). There was minimal staining of the stromal component. The hormonal status of these patients was not available.
Additional analysis on hormonal status was performed on the patients with cervical neoplasia and revealed that there was no link between p16INK4a expression in the cervical samples and day in menstrual cycle and/or exogenous hormone use.
Analysis of the recut slides versus the original diagnosis revealed substantial reproducibility of the diagnosis (weighted κ: 0.68 [95% CI = 0.58–0.72]). The correlation between p16INK4a and Ki-67 expression and neoplasia (CIN I or greater) was slightly better with the recut diagnosis versus the original diagnoses. Interobserver reproducibility in grading p16INK4a and Ki-67 immunostaining between a pathologist (SNA) and a technician (JM) was substantial (weighted κ = 0.74 [95% CI = 0.65–0.83] for p16INK4a and 0.70 [95% CI = 0.59–0.80] for Ki-67), whereas in determining p16INK4a negative or positive (≥1+ staining), agreement was excellent (Kappa = 0.84 [95% CI = 0.73–0.94]).
DISCUSSION
Because of the significant inter- and intraobserver variability in interpreting both Pap smears and cervical biopsy specimens, and the relatively low sensitivity of Pap smears, the search is on for better biomarkers to assist in the screening for and diagnosis of cervical neoplasia (1, 2). HPV testing by HybridCapture2, which is very sensitive for cervical neoplasia and useful in the triaging of ASCUS smears, seems to not be as specific for cervical neoplasia, as has been shown in other studies and in our study (2, 3). Many patients who are HPV positive do not have evidence of dysplasia by cytology or histology (2, 3). Recent research has analyzed the presence of p16INK4a in cervical neoplasia and has found a relationship between p16INK4a expression, high-risk HPV types, and cervical neoplasia, raising hope that p16INK4a could represent a specific and sensitive marker for cervical neoplasia (6, 8, 10).
In their study of Ki-67, cyclin E, and p16INK4a in 99 cervical biopsies, Keating and colleagues (10) found that 70% of HSIL lesions had diffuse strong expression of p16INK4a, whereas 23% had focal strong staining, similar to the findings in our study. As opposed to their finding of expression of p16INK4a in 100% of their CIN I cases, though, we found that only 57% of CIN I cases showed 1+ or stronger expression of p16INK4a (65% of the CIN I cases with the recut diagnosis). This is more in line with the findings by Klaes and colleagues who found that only 60% of their CIN I lesions had strong expression of p16INK4a, while 40% had only focal expression or none at all (6).
A possible reason for the lower expression of p16INK4a in low-grade lesions may be because a certain percentage are thought to be caused by low-risk HPV types (approximately 20% of low-grade lesions in the ASCUS/Low-grade Triage Study were negative for high-risk HPV types (19)). Because the affinity of the E7 protein of low-risk HPV for Rb is much lower than that of high-risk PHV types, there would not be overexpression of p16INK4a (20). Additionally, Keating et al. (10) show in their study that low-risk HPV is associated with less p16INK4a expression, and they suggest that different stages of high-risk HPV-induced cervical neoplasia may have different levels of p16INK4a expression.
Klaes et al. (6) showed in their study that 58 of 60 invasive cervical carcinomas expressed p16INK4a (52 of 53 [98%] squamous carcinomas and 6 of 7 [86%] adenocarcinomas), 5 of which were high-risk HPV negative. Our study had a slightly lower frequency of p16INK4a expression in invasive carcinomas (91% of squamous carcinomas and 71% of adenocarcinomas) and a lower frequency of HPV positivity by PCR (73%); none of the HPV-positive tumors were negative for p16INK4a expression. This slightly lower frequency of p16INK4a expression and lower rate HPV detection is probably secondary to a couple of factors: the tumors in our study came from African patients with very advanced-stage disease, and the samples were often extensively necrotic. (Decreased detection of HPV in advanced stage tumors is a known phenomenon) (21). In the study by Sano and colleagues (8), they found that 5 of 17 (29%) of their HPV negative cervical cancers lacked strong p16INK4a expression, and overall 17 of 54 (31%) of their cervical cancers lacked HPV by PCR. Interestingly, they also showed a lower frequency of p16INK4a expression in their adenocarcinomas, similar to the findings in our study and the study by Klaes and colleagues (6).
In atypical lesions, Keating and colleagues (10) found that p16INK4a positivity in lesions with squamous metaplastic atypia correlated significantly with the presence of high-risk HPV types. In our study, most of the p16INK4a positive atypical lesions were either associated with high-risk HPV types, had a significant history of dysplasia, or had concurrent dysplasia. Unfortunately, the number of atypical squamous metaplastic lesions in our study was small, and follow-up was lacking in some cases.
Although Ki-67 has been shown in several studies to be useful in diagnosing cervical neoplasia, our study shows that whereas Ki-67 expression is a sensitive marker of cervical neoplasia, overexpression is linked to both inflammation and reactive/atypical changes; p16INK4a showed no such association (7, 10, 14, 22). This is to be expected as inflammation is associated with increased cell turnover and reactive cervical squamous metaplasia has been shown to overexpress Ki-67 (10).
Also, although Ki-67 may be useful in the diagnosis of cervical neoplasia on histology, using Ki-67 as a potential screening marker is problematic as Ki-67 is expressed in all proliferating cells and thus is not a specific marker for cervical neoplasia or HPV infection, but rather an indirect marker of neoplasia. p16INK4a expression on the other hand is thought to be linked to HPV infection in cervical epithelium and specifically epithelium that has progressed in the neoplastic pathway sufficiently to allow for inactivation of Rb and overexpression of p16INK4a (6, 8). The utility of p16INK4a in screening for cervical neoplasia in cervicovaginal samples remains to be seen, but recent publications indicate that p16INK4a can detect cervical neoplasia in cytology samples (23).
Because p16INK4a has potential as a screening test, we thought to test consecutive endometrial samples, as the expression of p16INK4a in endometrial samples has been reported previously and endometrial cells are commonly present in cervicovaginal cytology samples (24, 25). In the study by Milde-Langosch and colleagues (24), they found that p16INK4a expression varied significantly with progesterone receptor status; however, we did not find any variability in p16INK4a expression according to the cycle status of the endometrium (secretory or proliferative). Shiozawa et al. (25) found p16INK4a staining in only the cytoplasm of proliferative endometrium and expression in 30% of their carcinoma samples. The expression of p16INK4a in our study was strong nuclear and cytoplasmic in the epithelium of the endometrium. However, as this was primarily a study of cervical neoplasia, only a limited number of cases were analyzed, and it is difficult to draw significant conclusions from this sample.
It is noteworthy that there was no significant variation of p16INK4a expression in cervical samples with the patient’s day in cycle or exogenous hormone use in our study, compared with the variation reported in the endometrium in the study by Milde-Langosch and colleagues (24).
The purpose of reviewing the recut specimens in our study was to confirm the continued presence of the lesion in the levels used for immunohistochemistry, as it has been shown that the diagnosis on cervical biopsies can vary significantly with levels (26), and to check for interobserver reproducibility. As shown above, the correlation between p16INK4a, Ki-67, and lesion grade improved with the recut slides, as is expected because many of the neoplastic lesions were small and were not present on all levels (i.e., “cut-through”). This was especially true for Ki-67 expression in CIN I lesions. When we analyzed p16INK4a expression versus the recut diagnosis almost all CIN III lesions expressed p16INK4a except one. This lesion was not without controversy too, as one of three pathologists categorized it as “atypical squamous metaplasia, cannot rule out high-grade neoplasia.”
As far as interobserver reproducibility, our study shows that p16INK4a immunohistochemistry interpretation is reproducible. The κ value between a pathologist and a technician in grading the immunohistochemistry ranged from substantial agreement to excellent agreement. This level of agreement is better than that found between pathologists in the ASCUS/Low-grade Triage Study on grading cervical neoplasia on biopsy specimens (1).
One of the strengths of this study is that it includes a large sample of women with negative, reactive, and atypical biopsies because the criteria for referral to colposcopy included women with negative Pap smears who were HPV DNA positive, in addition to a random sample of women with negative Pap and HPV DNA test results. Thus, our study had a large portion of women with positive HPV results who were negative by cytology and histology. Most studies of biopsy-confirmed cervical dysplasia contain few, if any, HPV positive, histologically and cytologically negative samples for comparison. Such a comparison group is critical for estimating the frequency of detecting specific biomarkers among those who do not have the disease of interest. If the observed frequency is low, it suggests that testing for the particular biomarker will have high specificity (i.e., a low number of false-positive test results) if used for screening the general population. The design of the study also made it possible to assess multiple biomarkers among a sample of reproductive-aged women from a clinical organization (Planned Parenthood) that provides a high percentage of Pap tests in this country. Women attending such clinics in different parts of the county appear to have similar risk profiles for HPV infection and for abnormal Pap tests, indicating that our results for p16INK4a and for Ki-67 may be generalizable to reproductive aged women throughout the country (19).
Some of the limitations of this study are that not all of the tests were performed on all of the specimens. However, consecutive specimens were tested and selection was unbiased as to test results. Furthermore, a better direct comparison of p16INK4a and Hybrid Capture2 or PCR for high-risk HPV DNA would include ELISA (enzyme-linked immunoassay) testing for p16INK4a on a cervicovaginal sample, but an ELISA test is not yet available.
As shown by this study, p16INK4a seems to be specific in the cervix for cervical neoplasia, especially high-grade cervical neoplasia, and seems to be more specific for neoplasia than Ki-67 expression. Though p16INK4a may help in the histological diagnosis of cervical neoplasia, it is not meant to be touted as a replacement for histology. p16INK4a may be more specific for cervical neoplasia than Hybrid Capture2 or PCR for high-risk HPV types, though this comparison is tenuous and further research with a liquid-based assay for p16INK4a is necessary to evaluate this biomarker’s usefulness in screening. The strong expression in endometrial epithelium might be problematic in testing cervicovaginal cytology samples (which is not an issue with Hybrid Capture2), but p16INK4a clearly has potential as a screening marker for cervical neoplasia.
References
Stoler MH, Schiffman M . Interobserver reproducibility of cervical cytologic and histologic interpretations: realistic estimates from the ASCUS-LSIL Triage Study. JAMA 2001; 285: 1500–1505.
Wright TC Jr, Denny L, Kuhn L, Pollack A, Lorincz A . HPV DNA testing of self-collected vaginal samples compared with cytologic screening to detect cervical cancer. JAMA 2000; 283: 81–86.
Manos MM, Kinney WK, Hurley LB, Sherman ME, Shieh-Ngai J, Kurman RJ, et al. Identifying women with cervical neoplasia: using human papillomavirus DNA testing for equivocal Papanicolaou results. JAMA 1999; 281: 1605–1610.
Lorincz AT, Reid R, Jenson AB, Greenberg MD, Lancaster W, Kurman RJ . Human papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types. Obstet Gynecol 1992; 79: 328–337.
Alani RM, Munger K . Human papillomaviruses and associated malignancies. J Clin Oncol 1998; 16: 330–337.
Klaes R, Friedrich T, Spitkovsky D, Ridder R, Rudy W, Petry U, et al. Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer 2001; 92: 276–284.
Keating JT, Ince T, Crum CP . Surrogate biomarkers of HPV infection in cervical neoplasia screening and diagnosis. Adv Anat Pathol 2001; 8: 83–92.
Sano T, Oyama T, Kashiwabara K, Fukuda T, Nakajima T . Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am J Pathol 1998; 153: 1741–1748.
Nakao Y, Yang X, Yokoyama M, Ferenczy A, Tang SC, Pater MM, et al. Induction of p16 during immortalization by HPV 16 and 18 and not during malignant transformation. Br J Cancer 1997; 75: 1410–1416.
Keating JT, Cviko A, Riethdorf S, Riethdorf L, Quade BJ, Sun D, et al. Ki-67, cyclin E, and p16INK4 are complimentary surrogate biomarkers for human papilloma virus-related cervical neoplasia. Am J Surg Pathol 2001; 25: 884–891.
Kulasingam SL, Hughes JP, Kiviat NB, Mao C, Weiss NS, Kuypers JM, et al. Evaluation of human papillomavirus testing in primary screening for cervical abnormalities: comparison of sensitivity, specificity, and frequency of referral. JAMA 2002; 288: 1749–1757.
Lin P, Koutsky LA, Critchlow CW, Apple RJ, Hawes SE, Hughes JP, et al. HLA class II DR-DQ and increased risk of cervical cancer among Senegalese women. Cancer Epidemiol Biomarkers Prev 2001; 10: 1037–1045.
Gown AM, deWever N, Battifora H . Microwave-based antigenic unmasking: a revolutionary new technique for routine immunohistochemistry. Appl Immunohistochem 1993; 1: 256–266.
Kruse AJ, Baak JP, de Bruin PC, Jiwa M, Snijders WP, Boodt PJ, et al. Ki-67 immunoquantitation in cervical intraepithelial neoplasia (CIN): a sensitive marker for grading. J Pathol 2001; 193: 48–54.
Sibony M, Commo F, Callard P, Gasc JM . Enhancement of mRNA in situ hybridization signal by microwave heating. Lab Invest 1995; 73: 586–591.
Sano T, Hikino T, Niwa Y, Kashiwabara K, Oyama T, Fukuda T, et al. In situ hybridization with biotinylated tyramide amplification: detection of human papillomavirus DNA in cervical neoplastic lesions. Mod Pathol 1998; 11: 19–23.
Gravitt PE, Peyton CL, Apple RJ, Wheeler CM . Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J Clin Microbiol 1998; 36: 3020–3027.
Peyton CL, Schiffman M, Lorincz AT, Hunt WC, Mielzynska I, Bratti C, et al. Comparison of PCR- and hybrid capture-based human papillomavirus detection systems using multiple cervical specimen collection strategies. J Clin Microbiol 1998; 36: 3248–3254.
Human papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intraepithelial lesions: baseline data from a randomized trial. The Atypical Squamous Cells of Undetermined Significance/Low-Grade Squamous Intraepithelial Lesions Triage Study (ALTS) Group. J Natl Cancer Inst 2000; 92: 397–402.
Gage JR, Meyers C, Wettstein FO . The E7 proteins of the nononcogenic human papillomavirus type 6b (HPV-6b) and of the oncogenic HPV-16 differ in retinoblastoma protein binding and other properties. J Virol 1990; 64: 723–730.
Jarrell MA, Heintz N, Howard P, Collins C, Badger G, Belinson J, et al. Squamous cell carcinoma of the cervix: HPV 16 and DNA ploidy as predictors of survival. Gynecol Oncol 1992; 46: 361–366.
al-Saleh W, Delvenne P, Greimers R, Fridman V, Doyen J, Boniver J . Assessment of Ki-67 antigen immunostaining in squamous intraepithelial lesions of the uterine cervix. Correlation with the histologic grade and human papillomavirus type. Am J Clin Pathol 1995; 104: 154–160.
Bibbo M, Klump WJ, DeCecco J, Kovatich AJ . Procedure for immunocytochemical detection of P16INK4A antigen in thin-layer, liquid-based specimens. Acta Cytol 2002; 46: 25–29.
Milde-Langosch K, Riethdorf L, Bamberger AM, Loning T . P16/MTS1 and pRB expression in endometrial carcinomas. Virchows Arch 1999; 434: 23–28.
Shiozawa T, Nikaido T, Shimizu M, Zhai Y, Fujii S . Immunohistochemical analysis of the expression of cdk4 and p16INK4 in human endometrioid-type endometrial carcinoma. Cancer 1997; 80: 2250–2256.
Luo VY, Prihoda TJ, Sharkey FE . Number of levels needed for diagnosis of cervical biopsies. Arch Pathol Lab Med 2002; 126: 1205–1208.
Acknowledgements
This work was supported by a grant from the National Institutes of Health (NCI CA34493).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Agoff, S., Lin, P., Morihara, J. et al. p16INK4a Expression Correlates with Degree of Cervical Neoplasia: A Comparison with Ki-67 Expression and Detection of High-Risk HPV Types. Mod Pathol 16, 665–673 (2003). https://doi.org/10.1097/01.MP.0000077518.78046.0C
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1097/01.MP.0000077518.78046.0C
Keywords
This article is cited by
-
Molecular landscape of vulvovaginal squamous cell carcinoma: new insights into molecular mechanisms of HPV-associated and HPV-independent squamous cell carcinoma
Modern Pathology (2022)
-
Liquid-based cytology: do ancillary techniques enhance detection of epithelial abnormalities?
Archives of Gynecology and Obstetrics (2018)
-
Introduction of p16INK4a as a surrogate biomarker for HPV in women with invasive cervical cancer in Sudan
Infectious Agents and Cancer (2017)
-
p16ink4 and cytokeratin 7 immunostaining in predicting HSIL outcome for low-grade squamous intraepithelial lesions: a case series, literature review and commentary
Modern Pathology (2016)
-
Increased RIPK4 expression is associated with progression and poor prognosis in cervical squamous cell carcinoma patients
Scientific Reports (2015)