Abstract
Immunohistochemistry (IHC) is used extensively to assess markers for prognosis and sensitivity to novel anticancer agents, as well as in the routine clinical assessment of cancers. Yet, although it is well known that tumors are highly heterogeneous, the resulting sampling error in the measurement of histological markers is often ignored, particularly in basic scientific studies. In this paper, we tested the hypothesis that the optimization of tissue sampling to compensate for heterogeneity improves the correlation between histological measurements of the intrinsic hypoxia marker carbonic anhydrase IX (CAIX) and global tumor oxygenation status. The study was based on a group of 24 patients with invasive cervical carcinoma from whom multiple biopsies were obtained at the time of direct pO2 assessment within the tumor, done as part of a research study. Measurements were made by image analysis of multiple deep sections cut through these biopsies, labeled for CAIX using both immunofluorescence and immunohistochemical techniques, and included tissue microarray (TMA) simulations. Variance and correlation analysis showed that the size of the tissue sample (biopsy or TMA core) was the major factor affecting accuracy of measurement in the sample. Sampling of multiple biopsies/cores also improved the global tumor assessment, provided that these were sufficiently separated in space. Optimization of sampling resulted in an improved correlation of CAIX staining with tumor pO2 measurements, consistent with the hypothesis. However, CAIX was inferior to pO2 measurements as a tool for patient stratification. Improved analytical methods to account for intratumoral heterogeneity are needed to provide reliable measurements of molecular markers.
Similar content being viewed by others
Main
The assessment of biological markers for prognostic, treatment and research purposes is increasingly important as new genes and proteins are identified as involved in specific disease states. Clinically important biomarkers such as estrogen/progesterone receptors and Her2-neu are extensively studied to evaluate sampling adequacy in biopsies and tissue microarrays (TMAs). Such assessment is, however, problematic for newly proposed biomarkers or markers that show heterogeneous spatial distribution because of inherent sampling error.1, 2, 3, 4, 5, 6, 7, 8 While such sampling error has been addressed in a number of publications particularly concerning TMAs,9, 10, 11, 12, 13, 14, 15, 16, 17, 18 the uncertainties of interpretation that it causes remain widely underappreciated, particularly in research studies. Following the work of a number of authors in animal models,19, 20, 21 we have investigated this issue in detail in relation to hypoxia in human cervical carcinomas.
Assessment of hypoxia has major prognostic, and potential therapeutic benefits, in a number of cancers.22, 23, 24, 25, 26 Because of its correlation with clinical outcome, the gold standard of assessing hypoxia is measurement of pO2 in vivo by probes like the Eppendorf polarographic electrode, which samples ∼100–150 points within the tumor. Although that technique provides assessment of global tumor state, it is cumbersome, invasive and not applicable to archival material. Reliable immunohistochemistry (IHC) techniques using putative endogeneous markers would make assessment of hypoxia easier and much more widely available. Carbonic anhydrase IX (CAIX), which is regulated by hypoxia-inducible transcription factors, is one such marker that has attracted attention due to its stability and the intense surface membrane staining typically seen in hypoxic tissue. Despite the wide use of CAIX as a hypoxia marker,27, 28, 29, 30, 31, 32 its expression level has not been proven to correlate with direct pO2 measurements. Initially, a moderate correlation was observed using a visual scoring system,33 but this was not confirmed in later reports utilizing both visual scoring and computerized image analysis.34, 35, 36 In a recent study of 110 patients with invasive cervical carcinoma done by our own group, we obtained multiple punch or core biopsies from a subgroup of patients. Measurements of CAIX expression in this limited amount of material suggested that intratumoral heterogeneity accounts for >40% of the total variance observed between tumors; a result that is consistent with studies done on spontaneous tumors in dogs using a nitroimidazole exogenous hypoxia marker.19, 20, 21 The present study was therefore designed to test the hypothesis that optimization of tissue sampling to compensate for intratumoral heterogeneity improves the correlation between CAIX staining and direct pO2 assessment.
MATERIALS AND METHODS
Patients
The study was approved by the University Health Network Research Ethics Board. Patient selection criteria, measurements of tumor oxygenation and tissue handling were performed as described previously.34, 37 Immediately following pO2 measurements, five punch biopsies per tumor were taken from different locations within the tumors of 30 patients. These locations were close to but not identical with the regions in which the pO2 measurements were made. We began by extensively sampling an initial series of 10 patients, using a fluorescence imaging technique similar to that previously described for CAIX measurements in cervical carcinomas.34 On the basis of the analysis of tumoral heterogeneity in this series, and a reevaluation of laboratory techniques, we then studied a total of 24 patients whose biopsies fulfilled the inclusion criteria to correlate CAIX with direct tumor pO2 measurements.
Initial Series
Cryostat sections were cut at 5-μm thickness. Multiple biopsies obtained from three of the patients were sectioned every 250 μm until the tissue was exhausted, to allow three-dimensional reconstruction. From the remaining seven tumors, four levels also spaced 250 μm apart were obtained. Biopsies were assessed by a pathologist (VI) during cutting to ensure >30% of tumor tissue. Sections were fixed in 2% formaldehyde/PBS for 20 min, incubated in primary antibody to CAIX (Novus biologicals, Littleton, CO, USA, rabbit polyclonal at 1:200) for 2 h and secondary antibody (anti-rabbit IgG Cy5 conjugated) for 1 h. The stained slides were imaged by fluorescence using a TISSUEscope laser scanning microscope (www.confocal.com Biomedical Photometrics Inc., Waterloo, Canada). This instrument is equipped with red, green and blue lasers, and can be used for fluorescent and transmitted light imaging. The slides were then re-stained using the hematoxylin–phloxin–saffron (HPS) trichrome technique and re-imaged by transmitted light using the same instrument settings. The green transmitted light was used for tumor–stroma separation by distinguishing the pink cytoplasm of tumor cells from the yellow stromal collagen. Fluorescence and transmission scanning were performed at 1 μm per pixel resolution and images were saved as eight-bit grayscale TIFF files.
Binary images were generated with Adobe® Photoshop® 7.0 (Adobe Systems Inc., USA). CAIX images were pasted as a layer to the corresponding HPS images, moved to match the tissue contours, and the bi-layered image cropped with the layers saved as separate files to produce aligned pairs of CAIX and HPS images. To avoid interobserver and intra-observer variation, thresholding to select positive staining was performed using a fixed value for the whole set. To compensate for occasional variations of tissue thickness, images with significantly darker CAIX negative areas were adjusted to match the dynamic range of the bright images. This technique was found superior to other normalization approaches, including manual image-by-image thresholding. Artifacts were manually erased by a pathologist (VI). A chosen threshold value was tested and adjusted on images with a range of CAIX positivity. This value was incorporated in a macro, and the images were batch processed. The HPS images were thresholded to create tumor masks in similar fashion. Each mask was assessed for accuracy by correlation with the histological slides and the original images. Signal-mask pairs were analyzed in batches using an image analysis program developed by one of the authors (AM; language IDL 6.3, ITT Visual Information Solutions, Boulder, CO, USA) and used in earlier studies.34, 38, 39 Tumoral area and percentage of CAIX-positive pixels in this area were obtained.
Immunofluorescence vs IHC
We investigated whether greater consistency in selecting positive staining could be achieved by using color images of IHC compared to grayscale imaging of immunofluorescence. Sixty consecutive 5 μm thick sections were cut from a SiHa cervical carcinoma xenograft, three sections per slide. The slides were stained in alternate order using immunofluorescence or an immunoperoxidase technique with diaminobenzidine as the chromogen. A total of 10 fluorescent and 10 immunoperoxidase labeled triplicates were analyzed by two observers independently (VI and AM).
Full Series Analysis
Sections were processed in one batch to reduce technical variables. Cryostat sections were fixed in 2% formaldehyde/PBS for 20 min, blocked for endogenous peroxidase and biotin, and incubated in primary antibody to CAIX (mouse monoclonal MN75 at 1:100) for 16 h. Immunoperoxidase staining was carried out using Idetect Ultra HRP detection system (ID Labs Inc., Canada) with diaminobenzidine as the substrate. The slides were scanned using a ScanScope CS (Aperio Technologies, CA, USA) at 20 × resolution and each section was saved as an eight-bit RGB TIFF file at 1 μm per pixel.
These images did not require normalization. A range of positive brown pixels was collected (Photoshop replace color tool), converted into black (zero intensity), tested on images representing a spectrum of CAIX positivity, and used for action recording. Stroma and artifacts were erased manually in the copies of the original images and a thresholding action separating tumor from the background was recorded. These actions were used for batch processing. Numerical data were collected as in the initial series, with the average value of triplicate sections calculated for each level. Additionally, an automated filling operation was applied to the brown signal images to calculate a ‘membranous staining/tumor area’ conversion ratio.
TMA Simulation
Images with white circles of 0.6 and 1.5 mm diameter representing TMA cores were pasted as a layer on the tumor masks to cover tumor area without the knowledge of CAIX distribution, as illustrated in Figure 1a. Separate sets of 1, 2 and 4 circles per biopsy were generated. For single cores the circle was placed in the middle, for two and four cores they were spaced evenly based on our previous studies with CAIX staining in soft-tissue sarcomas.40 Then the images were thresholded to select only tumor area within the circle. Using the same approach, tumor masks were additionally sampled three times by 0.6 mm circles, where circle 1 sampled tissue sections close to one edge, circle 2 immediately adjacent to circle 1 (0.6 mm between centers) and circle 3 was placed close to the edge opposite to circle 1 (3–5 mm apart).
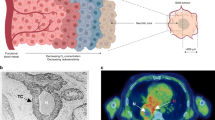
Intratumoral heterogeneity of carbonic anhydrase IX (CAIX) in the x–y and z axes. (a) Immunoperoxidase staining for CAIX in a single tissue section. Analysis of the entire section gave a value of 10.8% CAIX labeling. The circles limit the analysis to 0.6 mm simulated tissue microarray (TMA) cores, and show a wide range in CAIX (for publication purpose only, the image was digitally enhanced to better visualize CAIX areas). (b) Three-dimensional restoration of CAIX immunofluorescence staining based on 16 sections spaced 0.25 mm apart, generated by ‘etdips’ image processing software (http://clinicalcenter.nih.gov/cip/software/etdips/). Red color indicates CAIX-positive foci, gray external surface of the biopsy.
Statistical Analysis
Using variance component analysis, the total variance was partitioned in accordance with the different sources of variation. For the initial data set of 10 tumors, the variance was split into three parts: ‘between patients’, ‘between biopsies’ and ‘between sections’. The full series of 24 tumors had an additional ‘within level’ variability of triplicate sections. The proportion of the variance attributable to each of these sources was calculated. The percent CAIX staining was arcsine transformed to stabilize the variance of the residuals. The analysis was performed for the whole data set as well as for subsets defined based on the size of the tumor area in the section. The proportions of misclassification for two cutoff points (5 and 10) were calculated assuming that the arcsine transformation of the CAIX values have a normal distribution with the variances estimated using the variance component analysis. A more detailed description of the statistical methods used is given in the Appendix.
RESULTS
Initial Series and Protocol Design
Data collected in the initial set were used for protocol design, prior to proceeding to a larger set of patient samples. A total of 47 biopsies from 10 patients were used to obtain 318 sections, including three cases where sections spaced 250 μm apart were used to build three-dimensional reconstructions of CAIX distribution within the tissue. As illustrated in Figure 1, there was considerable heterogeneity in CAIX staining in all three planes.
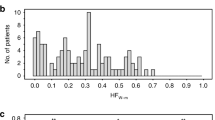
Since we limited CAIX measurement to the tumoral tissue, we assessed usable size of the biopsies by tumor area in a section. The median tumor area per section in this set was 4.9 mm2 (range 0.2–17.8 mm2). The median percent of CAIX-positive pixels within the tumor area was 5.63% (range 0.17–27.5%). Since CAIX staining has a membranous pattern, the area of staining measured by image analysis is smaller than the area occupied by the cells expressing CAIX visually estimated by observers. We calculated a conversion factor to compare our results with the studies estimating/scoring CAIX area visually. The unstained nuclei and cytoplasm were filled by an algorithm, and the ‘membranous staining/tumor area’ ratio was calculated. The median ratio was 1.7; for example, a value of 10% of CAIX-positive pixels was equivalent to about 17% of visually perceived tumor area (Supplementary Figure 1).
To design the sampling protocol for a larger set of samples, we performed a detailed variance analysis. When CAIX values measured within sections were arranged in the sequence in which the sections were cut (Figure 2), they formed an irregular waveform similar to that which we observed previously along the lengths of core needle biopsies.34 There was noticeably larger measurement variation within the smaller biopsies containing <4 mm2 of tumor area per section (Figure 3a).
Percentage of carbonic anhydrase IX (CAIX)-positive pixels within the three (a, b and c) tumors with fully sectioned biopsies. Each (a, b and c) panel is for one patient, five biopsies per patient, each point gives CAIX value within an individual section. The sections are shown in the sequence in which they were cut through the block. The variation within and between biopsies was apparent with different amplitude between biopsies and patients.
(a) Variance components within groups of biopsies of increasing tumor area. The analysis was repeated when only the sections with tumor area above a certain level were used (i.e. >0.5, ≥1 mm2, and so on). The variance ‘between patients’ reflects the biological difference between tumors, the variance ‘between biopsies’ reflects true intratumoral heterogeneity, and the variance ‘within biopsy’ includes the heterogeneity within biopsies as well as background technical variability (noise of measurement). Increasing biopsy size affected the ‘within’ and ‘between biopsy’ components, improving both the focal and the global tumor representation by a single section. Smaller biopsies had a larger difference between maximum and minimum values, with the smallest samples showing a range from near zero background to full saturation of the measured signal. (b) s.e. of CAIX measurement in combinations of biopsies and sections. Taking the same number of sections from different biopsies rather than the same biopsy is more effective reducing the error (compare for example the ‘1 biopsy—3 sections’ and ‘3 biopsies—1 section’ scenarios, which both provide three sections for analysis).
Variation within tumors accounted for 53% of the total variation (Figure 3a). The ‘between biopsies’ component reflected differences between parts of the tumor and the ‘within biopsy’ variation included both the technical error and the heterogeneity within biopsies. Individual sections of the larger biopsies were more representative of the whole biopsy and the whole tumor. The progressive improvement of accuracy was more pronounced within the smaller biopsy groups, with a trend to plateau with sections >4 mm2 tumor area. Since we could not measure tumor area during sectioning, we needed to determine an external diameter as a size cutoff. Sections containing 4 mm2 of tumor area had an average external diameter of 4 mm, which was accepted as the cutoff.
Figure 3b shows the effects of cumulative tissue sample size on the standard error of measurement, and indicates that increasing the number of biopsies reduce the error more effectively than increasing the number of sections within a biopsy. However, the additional requirement for biopsies ≥4 mm size and containing >70% tumor to minimize the variance (Figure 3a) limited the number of cases where adequate material was available for a three biopsy analysis. We therefore adopted a ‘2 biopsies—3 sections’ sampling protocol, in which two of the three levels were cut from the opposite sides of the frozen tissue blocks, to increase spatial separation while preserving the integrity of the samples (Table 1).
Fluorescence vs Immunohistochemical Staining
To choose a technique that gave greater reproducibility of CAIX measurement, immunofluorescent and immunohistochemical staining of xenograft material were compared by two observers independently. To assure that the techniques were measuring similar distributions and amounts of CAIX, sequential slices were stained by the two methods in alternate order (Supplementary Figure 2). The interobserver correlation was 0.93 for brown color images vs 0.67 for the fluorescent grayscale images. This finding was attributed to the sharper demarcation of positive–negative edges by color, whereas the grayscale fluorescent images had a gradual transition into the tissue autofluorescence background. We also compared the variation of CAIX values between serial sections cut at 5, 10 and 20 μm thickness, and found that 10 μm sections had significantly less section-to-section variation than 5 μm sections (variance=0.12 vs 0.5, respectively; not transformed value). The 20-μm sections showed patchy background staining due to reagent trapping, and were therefore unsuitable. On the basis of these considerations a final protocol was chosen, and this is summarized in Table 1.
Full Series
A total of 24 of the 30 patients fulfilled the criteria of having at least two biopsies of sufficient size and tumor content. As a result of this selection process, the median tumor area per section was 7.0 mm2, compared to 4.9 mm2 in the initial set. The median combined tumor area was 134 mm2 per patient (sum of triplicates for each level in two biopsies; total of 18 sections). Median CAIX value per section was 9.05% (range 0.08–57.9%) and 8.25% averaged per patient (range 0.46–37.25%). All data points are summarized in Figure 4. The variance analysis showed that the ‘within patient’ component decreased to 40% (from 53% in the initial series) showing that individual biopsies were more representative of the whole tumor. The variance between triplicate sections (within level), which reflects technical error, was 1.6% of total variance.
Percent of carbonic anhydrase IX (CAIX) staining values of the 24 patients (full series) arranged in increasing averaged value order; patients' data are numbered and separated by vertical grids, the three levels of each biopsy are connected by a line, each data point indicates a per-triplicate average value with the error bar showing low and high values within a level. Note the difference of values between the first and second level (250 μm apart) and the second and the third (cut from the opposite side, >3 mm).
To assess CAIX as a tool to categorize patients as normoxic/hypoxic, we calculated the 95% confidence interval (CI) for each patient (Figure 5a). In practice, categorization is achieved by selecting patients with a value above a chosen cutoff. The cutoff can be a median value of a data set or another value shown to be predictive of a reference parameter. Those patients whose 95% CI spans the cutoff value cannot be categorized with 95% confidence, forming a ‘gray zone’. For example, when a cutoff was set at CAIX value of 5%, 15 tumors (63%) could not be assigned to either group with 95% confidence. A cutoff set at zero value, categorizing tumors with any amount of detectable staining as hypoxic, produced the smallest ‘gray zone’ of six tumors (25%).
The 95% CI of the averaged per patient % carbonic anhydrase IX (CAIX) value, in full sections (a) and in tissue microarray (TMA) simulations (b and c). Solid black circles indicate averaged % CAIX staining of full sections, crosses—simulated microarray cores, dashed line—95% confidence limits, horizontal reference lines are drawn through 5 and 10% CAIX values. Single arrowheads indicate points beyond which hypoxic/normoxic stratification can be achieved with 95% confidence for a threshold drawn through 5% CAIX, and double arrowheads 15% CAIX. For example, for the full sections the width of 95% CI would not allow categorization of 15 tumors (63%) as hypoxic/normoxic when the cutoff is set at 5% (data points between single arrowheads) and 9 tumors (38%) when the cutoff is set at 15% (data points between double arrowheads) (a). The fraction of uncategorized tumors increases with reduction of the sampled tissue by microarray cores where a 15% threshold left 17 (70%) and 21 (88%) tumors in the ‘gray zone’ in the 8 (2 biopsies) and 4 (1 biopsy) core scenarios respectively (b and c). The smallest ‘gray zone’ is achieved by thresholding through zero value, which reduces the confidence corridor by the lower half (41% of tumors for simulated tissue microarray and 25% for the full sections).
As seen in Figure 4, the difference between the values of the first and second tissue section levels, which were spaced 250 μm apart, was significantly less (3% of the total variance) than that between the first and the third level, which was cut from the opposite side of the block (44% of the total within patient variance). This indicates that variation of CAIX measurement is greater in sections spaced further apart. In the initial series, we observed that the separation between the highest and lowest values within a biopsy could span over 1 mm (Figure 2). Insufficiently spaced levels may therefore sample either a focally marker-rich or depleted region, which affects estimation of the average value. Sampling from two levels taken from opposite sides of the biopsy appears practical to provide maximum spacing while preserving integrity of tissue.
Comparison with Direct Tumor pO2 Measurements
Similar to our CAIX data, direct pO2 measurement using the Eppendorf polarographic electrode, which was carried out in these patients, also involves multiple (∼100–150 points) measurements within the tumor. The results are presented as HP5, which is the percentage of pO2 values <5 mm Hg within the tumor. Figure 6a and b shows the 95% CI for each of these two sets of results. HP5 value is a product of a large number of point measurements, which results in a narrow CI. This, together with a large range of values between tumors, markedly reduces the gray zone categorizing into hypoxic/normoxic. The correlation between %CAIX and HP5 with a 2 × 2 table is shown in Figure 6c. Despite the optimized analytical and sampling protocol, the CAIX percentage values of the biopsies were only weakly correlated with the oxygenation status of the tumor measured using the Eppendorf polarographic electrode (Pearson=0.30, Spearman=0.35, P=0.091). When we excluded the outlier with a HP5 value of zero (Figure 6c), the correlation was more significant (Spearman=0.53, P=0.016). However, we were not able to exclude this patient sample as technically erroneous, and included it in the further analysis. The correlation between CAIX and HP5 varied significantly when we repeated this analysis using only one biopsy per tumor (Figure 7c). When we generated 2000 combinations, randomly using one out of two biopsies per tumor, the range of the Spearman's correlation coefficients was 0.039–0.55 (first and third quartile 0.24 and 0.39, respectively, median 0.32). Approximately 16% of these randomly generated values from one biopsy gave statistically significant (P<0.05) correlation coefficients between CAIX staining and the HP5 values. Observing this large range of correlation for average value, we tested if the maximum CAIX value correlates better with pO2 measurements. CAIX assessment was limited to a standardized area and the maximum values were used for correlation with HP5. The correlation coefficients were similar to that of the average values of two biopsies (Pearson=0.31, Spearman=0.38).
Comparison of 95% CI values for % carbonic anhydrase IX (CAIX) staining (a), and direct pO2 measurements (b). Data points indicate averaged % CAIX values within histological sections per patient and percentage of electrode pO2 measurements below 5 mm Hg. (c) shows the correlation, with the median values for each measurement indicated by dotted lines. Note that HP5 values, as a product of ∼100–150 point measurements, have narrow CI in addition to a larger range of values between tumors. These two factors markedly reduce the ‘gray zone’ of tumors which cannot be categorized with 95% confidence: two tumors (8%) have CI crossing median HP5 value in Figure 7b compared to 16 (67%) for CAIX in Figure 7a.
The effect of distance between TMA cores on estimation of the full section value (a) relationship of correlation between HP5 and sampling extent of one section (b), relationship of correlation with HP5 and distance between sections in a biopsy (c). (a) Empty circles indicate average values of two 0.6 mm cores sampling either adjacent (1&2) or remote (1&3) areas of the same section, carbonic anhydrase IX (CAIX) value of full section is accepted as 100%. Variance between two cores is shown for both scenarios (calculated on arcsine transformed data). Cores placed further apart had larger difference between their individual values (larger variance within pairs) producing averages closer to the value of the full section. (b) Empty circles indicate Spearman's correlation of data sets when only one section per tumor is analyzed and sections are sampled by increasing number and size of TMA cores; solid lines—median correlation of each sampling scenario; cross—the final correlation value of two biopsies. The number of cores had greater effect on correlation than the core diameter. For practical considerations, four cores per section placed in quadrants were equal to the full sections for correlation analysis. Note that sampling by one level per tumor can randomly have a large spread of correlation values. (c) Box and whisker plot (range, first and third quartiles and median) of CAIX correlation (Spearman) with HP5 in 2000 random combinations of sampling a tumor by one biopsy. Note that sections (levels) spaced further apart improve correlation. The range of correlation for sampling by one biopsy is large which shows the probability of obtaining contradicting results.
TMA Simulation
This was carried out to elaborate sampling protocols for TMAs, test accuracy of measurement of CAIX achievable with the TMA technique, and analyze heterogeneity within individual sections. As illustrated in Figure 5, we observed a direct relationship between the cumulative sample size and degree of confidence of patient stratification. Both the increase in the core diameter as well as the number of cores had a positive effect on measurement accuracy. The CAIX values of the full sections were used as a reference standard to study the effect of core spacing, illustrated in Figure 7. There was a greater difference between the values when cores were spaced further apart (cores 1&3, 3–5 mm apart, variance 0.019) than those sampling adjacent regions (cores 1&2, 0.6 mm apart, variance 0.009). Cores spaced further apart yielded average values closer to the value of the whole section (Figure 7a). To study the effect of core number and their size on correlation with pO2, we compared different sampling scenarios. Adding more cores was more effective in achieving better correlation than increasing the core diameter (Figure 7b). Four cores of either 0.6 or 1.5 mm sampling four quadrants produced a correlation similar to the full sections. These findings are consistent with our studies of CAIX expression in soft-tissue sarcomas.40
DISCUSSION
Expression of CAIX protein is greatly increased in response to hypoxia in cell lines and colocalizes to extrinsic nitroimidazole probes for hypoxia in tumor tissue, including samples obtained from cancer patients.41, 42, 43, 44, 45, 46, 47, 48 Early reports suggested that semiquantitative scoring of CAIX labeling was correlated with direct pO2 measurements in solid tumors, but this has not been confirmed by other groups.32, 33, 34, 35, 36, 49, 50, 51, 52, 53 When we examined a series of single punch biopsies obtained from 110 cervix cancer patients using automated image analysis, the percentage of tumoral area staining for CAIX was not correlated with direct pO2 measurements or with patient outcome.34 In a limited number of patients from that series we were able to measure CAIX levels in multiple punch biopsies, or along the lengths of core biopsies, and observed variance due to intratumoral heterogeneity that we estimated to represent >40% of the variance due to the difference of CAIX expression between individual patients. We therefore re-examined the problem by studying the effect of intratumoral heterogeneity of CAIX staining on the analytical methods used in clinical research: patient stratification and correlation with a relevant parameter. This is of particular interest because oxygen probe measurements are cumbersome and difficult to perform for many tumors. Furthermore, access to suitable probes for such measurements in patients is limited and simpler techniques to assess the extent of hypoxia in human tumors are needed.
Analysis of an initial set of samples showed that measurement accuracy of individual biopsies improves with increasing biopsy size (Figure 3a). Smaller biopsies had larger difference between their values, which is consistent with the statistical knowledge that maximum and minimum values are a function of sample size. To analyze strategies of sampling by multiple biopsies, we measured CAIX by extensive sampling aimed to produce reference values of global tumor CAIX. This allowed us to test which strategies can achieve values close to the reference with the least amount of sampling. Additional biopsies were more effective than additional sections as shown in Figure 3b. Therefore, we based our further analysis on the largest two biopsies per tumor. After we reduced the analytical error and utilized an elaborate sampling protocol, we found a weak correlation between the extent of CAIX expression and the Eppendorf probe readings. The degree of correlation had significant variation when only one biopsy per tumor was analyzed, which showed the effect of heterogeneity and the possibility of contradictory results due to random variability within undersampled material (Figure 7c).
The observed correlation is consistent with the idea that CAIX labeling identifies regions of tumor hypoxia. However, the distributional heterogeneity of CAIX produced a high sampling error that affected the degree of confidence of patient stratification. This is demonstrated by attempts to stratify the tumors using value cutoff values of CAIX staining. Over 40% of tumors could not be categorized with 95% confidence. For an accurate categorization of the tumors, the confidence interval needs to be much smaller than the true difference between the tumors, which is achievable by a large number of samples similarly to pO2 measurements (Figure 6). As a practical approach for the situations when adequate sampling is not achievable, thresholding through the zero value reduces the confidence corridor by the lower half and narrows the ‘gray zone’ of the uncategorized tumors (Figure 5). In other words, detection of any amount of staining is more accurate than quantification and thresholding through an above-zero value. However, this is not practicable in situations similar to CAIX assessment in cervical carcinomas, since virtually all of these express the protein to a different degree.34
Several factors need to be considered in circumstances when quantitative assessment of a marker is required. Ultimately, uniform sampling of the whole tumor would produce the best estimate of average or total value. For single biopsies, a larger size of tissue sample is needed to reduce the sampling error. For multiple biopsies (cores), larger numbers of well-spaced samples gives better representation of the whole tumor, while their size and correct placement add to their sampling value. Clustering of samples introduces bias by sampling the same focus and may result in reduced correlation (Figure 7c). As a practical finding, we observed that sections cut from opposite sides of a biopsy have similar range of values to that of different biopsies, and therefore have similar sampling value (Figure 7b and c).
We performed TMA simulations to test sampling protocols for TMAs, and study heterogeneity of CAIX in the x–y plane within individual sections. We observed larger difference between samples spaced further apart in the x–y plane during sampling of sections by three simulated cores. Cores spaced 0.6 and 3–5 mm within sections had variance 0.009 and 0.019, respectively (arcsine transformed data; Figure 7a), while full sections spaced 0.25 and 3–5 mm apart in the z axis showed variance of 0.0007 and 0.0067, where the smaller difference (variance) between full sections is due to the larger size of the tissue samples. Spacing samples further apart in all three dimensions increased the probability of sampling both marker-rich and depleted areas, and improved estimation of the average and correlation with pO2 status (Figure 7). These data are relevant only to estimation of average/median values. It needs to be emphasized that since variance is dependent on sample size, samples standardized by tumor volume can only be used to target maximum/minimum values.
Sampling error represents a general problem when assessing biological markers that show heterogeneous distribution in tumor tissue, in particular it likely explains some of the inconsistencies in the published literature regarding CAIX. Although several biomarkers have been used to validate sampling adequacy of small biopsies and TMAs, the data cannot be generalized due to a large range of distributional patterns and tumor architecture.9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 54 As we showed, random variability within undersampled material can give contradictory results, therefore results of sampling validation need to be reproduced by independent researchers for acceptance. Presently, there are no standardized tools to assess sampling error in biological tissues yet surprisingly little systematic work appears to have been reported in this area. With the increasing application of small biopsy samples and TMAs for the measurement of molecular markers relevant to cancer patient prognosis and treatment, further research into this problem is clearly needed.
References
Bachtiary B, Boutros PC, Pintilie M, et al. Gene expression profiling in cervical cancer: an exploration of intratumor heterogeneity. Clin Cancer Res 2006;12:5632–5640.
Ohmori H, Fujii K, Sasahira T, et al. Determinants for prediction of malignant potential by metalloproteinase:E-cadherin ratio in prostate core needle biopsy. Pathobiology 2006;73:98–104.
Tonotsuka N, Hosoi Y, Miyazaki S, et al. Heterogeneous expression of DNA-dependent protein kinase in esophageal cancer and normal epithelium. Int J Mol Med 2006;18:441–447.
Van Meter T, Dumur C, Hafez N, et al. Microarray analysis of MRI-defined tissue samples in glioblastoma reveals differences in regional expression of therapeutic targets. Diagn Mol Pathol 2006;15:195–205.
Kimura M, Tsuda H, Morita D, et al. Usefulness and limitation of multiple endoscopic biopsy sampling for epidermal growth factor receptor and c-erbB-2 testing in patients with gastric adenocarcinoma. Jpn J Clin Oncol 2005;35:324–331.
Chapman JA, Wolman E, Wolman SR, et al. Assessing genetic markers of tumour progression in the context of intratumour heterogeneity. Cytometry 1998;31:67–73.
Lewis JT, Ketterling RP, Halling KC, et al. Analysis of intratumoral heterogeneity and amplification status in breast carcinomas with equivocal (2+) HER-2 immunostaining. Am J Clin Pathol 2005;124:273–281.
Blackhall FH, Pintilie M, Wigle DA, et al. Stability and heterogeneity of expression profiles in lung cancer specimens harvested following surgical resection. Neoplasia 2004;6:761–767.
Pacifico MD, Grover R, Richman P, et al. Validation of tissue microarray for the immunohistochemical profiling of melanoma. Melanoma Res 2004;14:39–42.
Gomaa W, Ke Y, Fujii H, et al. Tissue microarray of head and neck squamous carcinoma: validation of the methodology for the study of cutaneous fatty acid-binding protein, vascular endothelial growth factor, involucrin and Ki-67. Virchows Arch 2005;447:701–709.
Singh SS, Qaqish B, Johnson JL, et al. Sampling strategy for prostate tissue microarrays for Ki-67 and androgen receptor biomarkers. Anal Quant Cytol Histol 2004;26:194–200.
Donati V, Faviana P, Dell'omodarme M, et al. Applications of tissue microarray technology in immunohistochemistry: a study on c-kit expression in small cell lung cancer. Hum Pathol 2004;35:1347–1352.
Gillett CE, Springall RJ, Barnes DM, et al. Multiple tissue core arrays in histopathology research: a validation study. J Pathol 2000;192:549–553.
Merseburger AS, Kuczyk MA, Serth J, et al. Limitations of tissue microarrays in the evaluation of focal alterations of bcl-2 and p53 in whole mount derived prostate tissues. Oncol Rep 2003;10:223–228.
Goethals L, Perneel C, Debucquoy A, et al. A new approach to the validation of tissue microarrays. J Pathol 2006;208:607–614.
Glockner S, Buurman H, Kleeberger W, et al. Marked intratumoral heterogeneity of c-myc and cyclinD1 but not of c-erbB2 amplification in breast cancer. Lab Invest 2002;82:1419–1426.
Chung GG, Zerkowski MP, Ghosh S, et al. Quantitative analysis of estrogen receptor heterogeneity in breast cancer. Lab Invest 2007;87:662–669.
Ruiz C, Seibt S, Al Kuraya K, et al. Tissue microarrays for comparing molecular features with proliferation activity in breast cancer. Int J Cancer 2006;118:2190–2194.
Thrall DE, Rosner GL, Azuma C, et al. Hypoxia marker labeling in tumor biopsies: quantification of labeling variation and criteria for biopsy sectioning. Radiother Oncol 1997;44:171–176.
Cline JM, Rosner GL, Raleigh JA, et al. Quantification of CCI-103F labeling heterogeneity in canine solid tumors. Int J Radiat Oncol Biol Phys 1997;37:655–662.
Cline JM, Thrall DE, Rosner GL, et al. Distribution of the hypoxia marker CCI-103F in canine tumors. Int J Radiat Oncol Biol Phys 1994;28:921–933.
Brizel DM, Scully SP, Harrelson JM, et al. Tumor oxygenation predicts for the likelihood of distant metastases in human soft tissue sarcoma. Cancer Res 1996;56:941–943.
Brizel DM, Sibley GS, Prosnitz LR, et al. Tumor hypoxia adversely affects the prognosis of carcinoma of the head and neck. Int J Radiat Oncol Biol Phys 1997;38:285–289.
Hockel M, Schlenger K, Aral B, et al. Association between tumor hypoxia and malignant progression in advanced cancer of the uterine cervix. Cancer Res 1996;56:4509–4515.
Hockel M, Vaupel P . Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst 2001;93:266–276.
Graeber TG, Osmanian C, Jacks T, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature 1996;379:88–91.
Nordsmark M, Loncaster J, Aquino-Parsons C, et al. Measurements of hypoxia using pimonidazole and polarographic oxygen-sensitive electrodes in human cervix carcinomas. Radiother Oncol 2003;67:35–44.
Nordsmark M, Loncaster J, Chou SC, et al. Invasive oxygen measurements and pimonidazole labeling in human cervix carcinoma. Int J Radiat Oncol Biol Phys 2001;49:581–586.
Maseide K, Kandel RA, Bell RS, et al. Carbonic anhydrase IX as a marker for poor prognosis in soft tissue sarcoma. Clin Cancer Res 2004;10:4464–4471.
Chia SK, Wykoff CC, Watson PH, et al. Prognostic significance of a novel hypoxia-regulated marker, carbonic anhydrase IX, in invasive breast carcinoma. J Clin Oncol 2001;19:3660–3668.
Hoskin PJ, Sibtain A, Daley FM, et al. GLUT1 and CAIX as intrinsic markers of hypoxia in bladder cancer: relationship with vascularity and proliferation as predictors of outcome of ARCON. Br J Cancer 2003;89:1290–1297.
Vordermark D, Brown JM . Endogenous markers of tumor hypoxia predictors of clinical radiation resistance? Strahlenther Onkol 2003;179:801–811.
Loncaster JA, Harris AL, Davidson SE, et al. Carbonic anhydrase (CA IX) expression, a potential new intrinsic marker of hypoxia: Correlations with tumor oxygen measurements and prognosis in locally advanced carcinoma of the cervix. Cancer Res 2001;61:6394–6399.
Hedley D, Pintilie M, Woo J, et al. Carbonic anhydrase IX expression, hypoxia, and prognosis in patients with uterine cervical carcinomas. Clin Cancer Res 2003;9:5666–5674.
Mayer A, Hockel M, Vaupel P . Carbonic anhydrase IX expression and tumor oxygenation status do not correlate at the microregional level in locally advanced cancers of the uterine cervix. Clin Cancer Res 2005;11:7220–7225.
Jankovic B, Aquino-Parsons C, Raleigh JA, et al. Comparison between pimonidazole binding, oxygen electrode measurements, and expression of endogenous hypoxia markers in cancer of the uterine cervix. Cytometry B Clin Cytom 2006;70:45–55.
Fyles A, Milosevic M, Hedley D, et al. Tumor hypoxia has independent predictor impact only in patients with node-negative cervix cancer. J Clin Oncol 2002;20:680–687.
Hedley D, Pintilie M, Woo J, et al. Up-regulation of the redox mediators thioredoxin and apurinic/apyrimidinic excision (APE)/Ref-1 in hypoxic microregions of invasive cervical carcinomas, mapped using multispectral, wide-field fluorescence image analysis. Am J Pathol 2004;164:557–565.
Vukovic V, Haugland HK, Nicklee T, et al. Hypoxia-inducible factor-1alpha is an intrinsic marker for hypoxia in cervical cancer xenografts. Cancer Res 2001;61:7394–7398.
Måseide K, Pintilie M, Kandel R, et al. Can sparsely and heterogeneously expressed proteins be detected using tissue microarrays? A simulation study of the hypoxia marker carbonic anhydrase IX (CA IX) in human soft tissue sarcoma. Pathology Research and Practice 2007, (in press).
Airley RE, Loncaster J, Raleigh JA, et al. GLUT-1 and CAIX as intrinsic markers of hypoxia in carcinoma of the cervix: relationship to pimonidazole binding. Int J Cancer 2003;104:85–91.
Troost EG, Bussink J, Kaanders JH, et al. Comparison of different methods of CAIX quantification in relation to hypoxia in three human head and neck tumor lines. Radiother Oncol 2005;76:194–199.
Goethals L, Debucquoy A, Perneel C, et al. Hypoxia in human colorectal adenocarcinoma: comparison between extrinsic and potential intrinsic hypoxia markers. Int J Radiat Oncol Biol Phys 2006;65:246–254.
Dubois L, Landuyt W, Haustermans K, et al. Evaluation of hypoxia in an experimental rat tumour model by [(18)F]fluoromisonidazole PET and immunohistochemistry. Br J Cancer 2004;91:1947–1954.
Beasley NJ, Wykoff CC, Watson PH, et al. Carbonic anhydrase IX, an endogenous hypoxia marker, expression in head and neck squamous cell carcinoma and its relationship to hypoxia, necrosis, and microvessel density. Cancer Res 2001;61:5262–5267.
Wykoff CC, Beasley NJ, Watson PH, et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res 2000;60:7075–7083.
Sobhanifar S, Aquino-Parsons C, Stanbridge EJ, et al. Reduced expression of hypoxia-inducible factor-1alpha in perinecrotic regions of solid tumors. Cancer Res 2005;65:7259–7266.
Olive PL, Aquino-Parsons C, MacPhail SH, et al. Carbonic anhydrase 9 as an endogenous marker for hypoxic cells in cervical cancer. Cancer Res 2001;61:8924–8929.
Olive PL, Banath JP, Aquino-Parsons C . Measuring hypoxia in solid tumours—is there a gold standard? Acta Oncol 2001;40:917–923.
Mayer A, Hockel M, Vaupel P . Endogenous hypoxia markers in locally advanced cancers of the uterine cervix: reality or wishful thinking? Strahlenther Onkol 2006;182:501–510.
Le QT, Chen E, Salim A, et al. An evaluation of tumor oxygenation and gene expression in patients with early stage non-small cell lung cancers. Clin Cancer Res 2006;12:1507–1514.
Mayer A, Hockel M, Vaupel P . Carbonic anhydrase IX expression and tumor oxygenation status do not correlate at the microregional level in locally advanced cancers of the uterine cervix. Clin Cancer Res 2005;11:7220–7225.
Nordsmark M, Loncaster J, Aquino-Parsons C, et al. The prognostic value of pimonidazole and tumour pO2 in human cervix carcinomas after radiation therapy: a prospective international multi-center study. Radiother Oncol 2006;80:123–131.
Ruiz C, Seibt S, Al Kuraya K, et al. Tissue microarrays for comparing molecular features with proliferation activity in breast cancer. Int J Cancer 2006;118:2190–2194.
Acknowledgements
VI was financially supported by a CHIR molecular oncologic pathology fellowship. Funding was also obtained from the NCI(C) program project grant raised by the Terry Fox Run.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website (http://www.laboratoryinvestigation.org)
Supplementary information
Appendix
Appendix
To stabilize the variance of the residuals the carbonic anhydrase IX (CAIX) values were first arcsine transformed:

where CAIX is the original proportion CAIX value and caix is the transformed value.
The average value and the 95% confidence intervals (CIs) per patient were calculated on the transformed values, caix and then transformed back to the original scale.
For each patient the caix value was calculated as the average of all values: two biopsies, three sections and three repeats. The 95% CI was calculated using the variance estimates obtained in the variance component analysis. Suppose that VB denotes the variance between biopsies, VS denotes the variance between sections and VR is the variance within a section. Then the variance of the average value CAIX is calculated as

The 95% CIs for caix are given by:

where zα is the α-quantile of the standard normal distribution. In this case since  . The last step is to transform back these values. Thus, for the average, the plotted value is
. The last step is to transform back these values. Thus, for the average, the plotted value is

and the limits are given by

The value HP5 per patient is defined as the proportion of measurements with Eppendorf value <5 mm Hg. The 95% CI for each patient was calculated using the normal approximation of the binomial distribution with the exception of one patient for which the exact CI was calculated. This patient had HP5=0. All the rest of the 23 patients had the condition n × p>10 fulfilled.
Rights and permissions
About this article
Cite this article
Iakovlev, V., Pintilie, M., Morrison, A. et al. Effect of distributional heterogeneity on the analysis of tumor hypoxia based on carbonic anhydrase IX. Lab Invest 87, 1206–1217 (2007). https://doi.org/10.1038/labinvest.3700680
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3700680
Keywords
This article is cited by
-
The role of proteomics in the age of immunotherapies
Mammalian Genome (2018)
-
Larger core size has superior technical and analytical accuracy in bladder tissue microarray
Laboratory Investigation (2017)
-
Downregulation of EGFR in hypoxic, diffusion-limited areas of squamous cell carcinomas of the head and neck
British Journal of Cancer (2016)
-
Analysis of the intra- and intertumoral heterogeneity of hypoxia in pancreatic cancer patients receiving the nitroimidazole tracer pimonidazole
British Journal of Cancer (2015)
-
B7-H3 expression in colorectal cancer: associations with clinicopathological parameters and patient outcome
BMC Cancer (2014)