Abstract
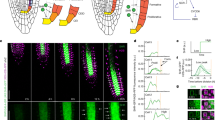
Positional information is pivotal for establishing developmental patterning in plants1,2,3, but little is known about the underlying signalling mechanisms. The Arabidopsis root radial pattern is generated through stereotyped division of initial cells and the subsequent acquisition of cell fate4. short-root (shr) mutants do not undergo the longitudinal cell division of the cortex/endodermis initial daughter cell, resulting in a single cell layer with only cortex attributes5,6. Thus, SHR is necessary for both cell division and endodermis specification5,6. SHR messenger RNA is found exclusively in the stele cells internal to the endodermis and cortex, indicating that it has a non-cell-autonomous mode of action6. Here we show that the SHR protein, a putative transcription factor, moves from the stele to a single layer of adjacent cells, where it enters the nucleus. Ectopic expression of SHR driven by the promoter of the downstream gene SCARECROW (SCR) results in autocatalytic reinforcement of SHR signalling, producing altered cell fates and multiplication of cell layers. These results support a model in which SHR protein acts both as a signal from the stele and as an activator of endodermal cell fate and SCR-mediated cell division.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
van den Berg, C., Willemsen, V., Hage, W., Weisbeek, P. & Scheres, B. Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378, 62–65 (1995).
van den Berg, C., Willemsen, V., Hendriks, G., Weisbeek, P. & Scheres, B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390, 287–289 (1997).
Kidner, C., Sundaresan, V., Roberts, K. & Dolan, L. Clonal analysis of the Arabidopsis root confirms that position, not lineage, determines cell fate. Planta 211, 191–199 (2000).
Dolan, L. et al. Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84 (1993).
Benfey, P. N. et al. Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis. Development 119, 57–70 (1993).
Helariutta, Y. et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567 (2000).
Di Laurenzio, L. et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433 (1996).
Knox, J. P., Linstead, P. J., King, J., Cooper, C. & Roberts, K. Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apieces. Planta 181, 512–521 (1990).
Freshour, G. et al. Developmental and tissue-specific structural alterations of the cell wall polysaccharides of Arabidopsis thaliana roots. Plant Physiology 110, 1413–1429 (1996).
Esau, K. Anatomy of Seed Plants 215–242 (Wiley, New York, 1977).
Malamy, J. & Benfey, P. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44 (1997).
Masucci, J. D. et al. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260 (1996).
Sabatini, S. et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472 (1999).
Lucas, W. et al. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270, 1980–1983 (1995).
Perbal, M., Haughn, G., Saedler, H. & Schwarz-Sommer, Z. Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development 122, 3433–3441 (1996).
Sessions, A., Yanofsky, M. F. & Weigel, D. Cell–cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 287, 419–421 (2000).
Jackson, D., Veit, B. & Hake, S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120, 405–413 (1994).
von Arnim, A. G., Deng, X.-W. & Stacey, M. G. Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221, 35–43 (1998).
Clough, S. J. & Bent, A. F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Wysocka-Diller, J., Helariutta, Y., Fukaki, H., Malamy, J. & Benfey, P. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127, 595–603 (2000).
Acknowledgements
We thank K. Roberts for the JIM13 antibody; M. Hahn for the CCRC-M2 antibody; B. Scheres for the QC46 marker line; J. Schiefelbein for the GL2::GUS line; A. von Arnim for the GFP plasmid; M. Aida for the pBIH vector; and M. Starz for the assistance with confocal microscopy. Multi-photon confocal images were taken with assistance by J. Feijo and N. Moreno. K.N. was supported by a fellowship from Japan Society for the Promotion of Science. This work was supported by a grant to P.N.B. from the NIH.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Video 1
Three-dimensional reconstruction of the root tip of the SHR::GFP fusion protein line, obtained through sequential longitudinal multiphoton optical sections.
Rights and permissions
About this article
Cite this article
Nakajima, K., Sena, G., Nawy, T. et al. Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307–311 (2001). https://doi.org/10.1038/35095061
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35095061
This article is cited by
-
BpWOX11 promotes adventitious root formation in Betula pendula
BMC Plant Biology (2024)
-
SHR and SCR coordinate root patterning and growth early in the cell cycle
Nature (2024)
-
BRIP1 and BRIP2 maintain root meristem by affecting auxin-mediated regulation
Planta (2024)
-
Identification and functional analysis of a CbSHR homolog in controlling adventitious root development in Catalpa bungei
Plant Cell, Tissue and Organ Culture (PCTOC) (2024)
-
Spatial distribution of three ARGONAUTEs regulates the anther phasiRNA pathway
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.