Featured
-
-
Article |
 In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target
In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target
In vivo CRISPR screening reveals that loss of Ptpn2 increases the response of tumour cells to immunotherapy and increases IFNγ signalling, suggesting that PTPN2 inhibition may potentiate the effect of immunotherapies that invoke an IFNγ response.
- Robert T. Manguso
- , Hans W. Pope
- & W. Nicholas Haining
-
Article |
 An atlas of human long non-coding RNAs with accurate 5′ ends
An atlas of human long non-coding RNAs with accurate 5′ ends
A catalogue of human long non-coding RNA genes and their expression profiles across samples from major human primary cell types, tissues and cell lines.
- Chung-Chau Hon
- , Jordan A. Ramilowski
- & Alistair R. R. Forrest
-
Letter |
 Local regulation of gene expression by lncRNA promoters, transcription and splicing
Local regulation of gene expression by lncRNA promoters, transcription and splicing
Various cis-regulatory functions of genomic loci that produce long non-coding RNAs are revealed, including instances where their promoters have enhancer-like activity and the lncRNA transcripts themselves are not required for activity.
- Jesse M. Engreitz
- , Jenna E. Haines
- & Eric S. Lander
-
Article |
 High-throughput discovery of novel developmental phenotypes
High-throughput discovery of novel developmental phenotypes
Identification and characterization, using a comprehensive embryonic phenotyping pipeline, of 410 lethal alleles during the generation of the first 1,751 of 5,000 unique gene knockouts produced by the International Mouse Phenotyping Consortium.
- Mary E. Dickinson
- , Ann M. Flenniken
- & Stephen A. Murray
-
Letter |
 Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens
Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens
A CRISPR screening approach shows that endoplasmic reticulum (ER)-associated protein complexes, including the oligosaccharyltransferase (OST) protein complex, are important for infection by dengue virus and other related mosquito-borne flaviviruses, whereas hepatitis C virus is dependent on distinct entry factors, RNA binding proteins and FAD biosynthesis.
- Caleb D. Marceau
- , Andreas S. Puschnik
- & Jan E. Carette
-
Article |
 Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice
Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice
PRDM9 is a DNA-binding protein that controls the position of double-strand breaks in meiosis, and the gene that encodes it is responsible for hybrid infertility between closely related mouse species; this hybrid infertility is eliminated by introducing the zinc-finger domain sequence from the human version of the PRDM9 gene, a change which alters both the position of double-strand breaks and the symmetry of PRDM9 binding and suggests that PRDM9 may have a more general but transient role in the early stages of speciation.
- Benjamin Davies
- , Edouard Hatton
- & Peter Donnelly
-
Letter |
 The spliceosome is a therapeutic vulnerability in MYC-driven cancer
The spliceosome is a therapeutic vulnerability in MYC-driven cancer
Splicing factors such as BUD31 are identified in a synthetic-lethal screen with cells overexpressing the transcription factor MYC; oncogenic MYC leads to an increase in pre-mRNA synthesis, and spliceosome inhibition impairs the growth and tumorigenicity of MYC-dependent breast cancers, suggesting that spliceosome components may be potential therapeutic targets for MYC-driven cancers.
- Tiffany Y.-T. Hsu
- , Lukas M. Simon
- & Thomas F. Westbrook
-
Letter |
 Integrator mediates the biogenesis of enhancer RNAs
Integrator mediates the biogenesis of enhancer RNAs
This study demonstrates a role for the Integrator complex in the stimulus-dependent induction of eRNAs and their 3′ processing; together with previously known roles of Integrator in transcription elongation and RNA processing, these results indicate that Integrator has broad functions in the regulation of eukaryotic gene expression.
- Fan Lai
- , Alessandro Gardini
- & Ramin Shiekhattar
-
Letter |
 Identification of cis-suppression of human disease mutations by comparative genomics
Identification of cis-suppression of human disease mutations by comparative genomics
Patterns of amino acid conservation have been used to guide the interpretation of the disease-causing potential of genetic variants in patients; now, an appreciable fraction of pathogenic alleles are shown to be fixed in the genomes of other species, suggesting that the genomic context has an important role in allele pathogenicity.
- Daniel M. Jordan
- , Stephan G. Frangakis
- & Nicholas Katsanis
-
Letter |
 Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation
Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation
The core promoters of developmental and housekeeping genes are shown to have distinct specificities for different enhancer sequences in Drosophila, and this specificity separates developmental and housekeeping gene regulatory programs across the genome.
- Muhammad A. Zabidi
- , Cosmas D. Arnold
- & Alexander Stark
-
Article
| Open Access Principles of regulatory information conservation between mouse and human
Principles of regulatory information conservation between mouse and human
As part of the mouse ENCODE project, genome-wide transcription factor (TF) occupancy repertoires and co-association patterns in mice and humans are studied; many aspects are conserved but the extent to which orthologous DNA segments are bound by TFs in mice and humans varies both among TFs and genomic location, and TF-occupied sequences whose occupancy is conserved tend to be pleiotropic and enriched for single nucleotide variants with known regulatory potential.
- Yong Cheng
- , Zhihai Ma
- & Michael P. Snyder
-
Letter |
 Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq
Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq
Single-cell transcriptome analysis enables the direct measurement of cell types and lineage hierarchies of the developing distal lung epithelium and identifies a population of bipotential alveolar progenitor cells.
- Barbara Treutlein
- , Doug G. Brownfield
- & Stephen R. Quake
-
Letter |
 High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells
High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells
This study describes the construction of a focused CRISPR/Cas-based lentiviral library in human cells and a method of gene identification based on functional screening and high-throughput sequencing analysis.
- Yuexin Zhou
- , Shiyou Zhu
- & Wensheng Wei
-
Article |
 Homologue engagement controls meiotic DNA break number and distribution
Homologue engagement controls meiotic DNA break number and distribution
DNA double-stranded breaks (DSBs) are shown to form in greater numbers in yeast cells lacking ZMM proteins, which are traditionally regarded as acting strictly downstream of DSB formation; these findings shed light on how cells balance the beneficial and deleterious outcomes of DSB formation.
- Drew Thacker
- , Neeman Mohibullah
- & Scott Keeney
-
Article
| Open Access Diversity and dynamics of the Drosophila transcriptome
Diversity and dynamics of the Drosophila transcriptome
A large-scale transcriptome analysis in Drosophila melanogaster, across tissues, cell types and conditions, provides insights into global patterns and diversity of transcription initiation, splicing, polyadenylation and non-coding RNA expression.
- James B. Brown
- , Nathan Boley
- & Susan E. Celniker
-
Article |
 The evolution of lncRNA repertoires and expression patterns in tetrapods
The evolution of lncRNA repertoires and expression patterns in tetrapods
Evolutionary study of long noncoding RNA (lncRNA) repertoires and expression patterns in 11 tetrapod species identifies approximately 11,000 primate-specific lncRNAs and 2,500 highly conserved lncRNAs, including approximately 400 genes that are likely to have ancient origins; many lncRNAs, particularly ancient ones, are actively regulated and may function mainly in embryonic development.
- Anamaria Necsulea
- , Magali Soumillon
- & Henrik Kaessmann
-
Letter |
 Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico
Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico
A risk haplotype for type 2 diabetes is identified with four amino acid substitutions in SLC16A11, which is present at ∼50% frequency in Native American samples and ∼10% in east Asian samples, but is rare in European and African samples; SLC16A11 may alter hepatic lipid metabolism, causing an increase in triacylglycerol levels.
- Amy L. Williams
- , Suzanne B. R. Jacobs
- & Teresa Tusié-Luna
-
Analysis |
 Inconsistency in large pharmacogenomic studies
Inconsistency in large pharmacogenomic studies
This Analysis compares two large-scale pharmacogenomic data sets that catalogued the sensitivity of a large number of cancer cell lines to approved and potential drugs, and finds that whereas the gene expression data are largely concordant between the two studies, the reported drug sensitivity measures and subsequently their association with genomic features are highly discordant.
- Benjamin Haibe-Kains
- , Nehme El-Hachem
- & John Quackenbush
-
Article |
 Transcriptome and genome sequencing uncovers functional variation in humans
Transcriptome and genome sequencing uncovers functional variation in humans
Sequencing and deep analysis of mRNA and miRNA from lymphoblastoid cell lines of 462 individuals from the 1000 Genomes Project reveal widespread genetic variation affecting the regulation of most genes, with transcript structure and expression level variation being equally common but genetically largely independent, and the analyses point to putative causal variants for dozens of disease-associated loci.
- Tuuli Lappalainen
- , Michael Sammeth
- & Emmanouil T. Dermitzakis
-
Article |
 RNAi screens in mice identify physiological regulators of oncogenic growth
RNAi screens in mice identify physiological regulators of oncogenic growth
Here, the first genome-wide in vivo RNA interference screens in a mammalian animal model are reported: genes involved in normal and abnormal epithelial cell growth are studied in developing skin tissue in mouse embryos, and among the findings, β-catenin is shown to act as an antagonist to normal epithelial cell growth as well as promoting oncogene-driven growth.
- Slobodan Beronja
- , Peter Janki
- & Elaine Fuchs
-
News & Views |
The inner life of proteins
A quantitative analysis shows that epistasis — the fact that genetic background determines whether a mutation is beneficial, deleterious or inconsequential — is the main factor regulating evolution at the level of proteins. See Letter p.535
- Günter P. Wagner
-
Article
| Open Access A physical, genetic and functional sequence assembly of the barley genome
A physical, genetic and functional sequence assembly of the barley genome
An integrated high-resolution genetic, physical and shotgun sequence assembly of the barley genome, one of the earliest domesticated and most important crops, is described; it will provide a platform for genome-assisted research and future crop improvement.
- Klaus F. X. Mayer
- , Robbie Waugh
- & Nils Stein
-
Letter |
Epistasis as the primary factor in molecular evolution
A comparison of more than 1,000 orthologues of diverse proteins shows that the rate of amino-acid substitution in recent evolution is an order of magnitude lower than that expected in the absence of epistasis, indicating that epistasis is pervasive throughout protein evolution.
- Michael S. Breen
- , Carsten Kemena
- & Fyodor A. Kondrashov
-
Article
| Open Access A map of rice genome variation reveals the origin of cultivated rice
A map of rice genome variation reveals the origin of cultivated rice
Whole-genome sequences of wild rice and cultivated rice varieties are used to produce a map of rice genome variation, and show that rice was probably first domesticated in southern China.
- Xuehui Huang
- , Nori Kurata
- & Bin Han
-
Letter |
 In vivo genome editing using a high-efficiency TALEN system
In vivo genome editing using a high-efficiency TALEN system
Although zebrafish is an important animal model for basic vertebrate biology and human disease modelling, rapid targeted genome modification has not been possible in this species; here a technique based on improved artificial transcription activator-like effector nucleases (TALENs) allows precise sequence modifications at pre-determined genomic locations.
- Victoria M. Bedell
- , Ying Wang
- & Stephen C. Ekker
-
Letter |
 Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2
Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2
Genetic analysis of breakthrough infections in people vaccinated against HIV-1 show that vaccine efficacy increased by up to 80% against viruses carrying two mutations in Env V2, but also raises the possibility of population-level adaptation to the vaccine.
- Morgane Rolland
- , Paul T. Edlefsen
- & Jerome H. Kim
-
-
-
Comment |
 Lessons for big-data projects
Lessons for big-data projects
To be successful, consortia need clear management, codes of conduct and participants who are committed to working for the common good, says ENCODE lead analysis coordinator Ewan Birney.
- Ewan Birney
-
News & Views Forum |
 ENCODE explained
ENCODE explained
The Encyclopedia of DNA Elements (ENCODE) project dishes up a hearty banquet of data that illuminate the roles of the functional elements of the human genome. Here, six scientists describe the project and discuss how the data are influencing research directions across many fields. See Articles p.57, p.75, p.83, p.91, p.101 & Letter p.109
- Joseph R. Ecker
- , Wendy A. Bickmore
- & Eran Segal
-
News Feature |
 ENCODE: The human encyclopaedia
ENCODE: The human encyclopaedia
First they sequenced it. Now they have surveyed its hinterlands. But no one knows how much more information the human genome holds, or when to stop looking for it.
- Brendan Maher
-
Article
| Open Access An integrated encyclopedia of DNA elements in the human genome
An integrated encyclopedia of DNA elements in the human genome
This overview of the ENCODE project outlines the data accumulated so far, revealing that 80% of the human genome now has at least one biochemical function assigned to it; the newly identified functional elements should aid the interpretation of results of genome-wide association studies, as many correspond to sites of association with human disease.
- Ian Dunham
- , Anshul Kundaje
- & Ewan Birney
-
Letter
| Open Access The long-range interaction landscape of gene promoters
The long-range interaction landscape of gene promoters
Chromosome conformation capture carbon copy (5C) is used to look at the relationships between functional elements and distal target genes in 1% of the human genome in three dimensions; the study describes numerous long-range interactions between promoters and distal sites that include elements resembling enhancers, promoters and CTCF-bound sites, their genomic distribution and complex interactions.
- Amartya Sanyal
- , Bryan R. Lajoie
- & Job Dekker
-
Letter |
 Genetic recombination is directed away from functional genomic elements in mice
Genetic recombination is directed away from functional genomic elements in mice
Comparison of Prdm9−/− and wild-type mice reveals a role for the PRDM9 protein in directing the recombination machinery away from important genomic regions.
- Kevin Brick
- , Fatima Smagulova
- & Galina V. Petukhova
-
Technology Feature |
 The changes that count
The changes that count
As more mutations are found across the genome, geneticists are focusing on learning which ones are likely to cause human disease, and how.
- Monya Baker
-
News |
 Video animation: RNA interference
Video animation: RNA interference
A video explaining RNA interference from Nature Reviews Genetics.
-
Letter |
 Predicting mutation outcome from early stochastic variation in genetic interaction partners
Predicting mutation outcome from early stochastic variation in genetic interaction partners
Mutations sometimes only affect a subset of genetically identical individuals; here, variation in the expression of chaperones and gene duplicates is shown to predict mutation outcome in C. elegans.
- Alejandro Burga
- , M. Olivia Casanueva
- & Ben Lehner
-
Article |
 New gene functions in megakaryopoiesis and platelet formation
New gene functions in megakaryopoiesis and platelet formation
A meta-analysis of genome-wide association studies in more than 66,000 individuals identifies 68 new genomic loci that reliably associate with platelet count and volume, and reveals new gene functions.
- Christian Gieger
- , Aparna Radhakrishnan
- & Nicole Soranzo
-
Article |
 Spatio-temporal transcriptome of the human brain
Spatio-temporal transcriptome of the human brain
- Hyo Jung Kang
- , Yuka Imamura Kawasawa
- & Nenad Šestan
-
Letter |
 Temporal dynamics and genetic control of transcription in the human prefrontal cortex
Temporal dynamics and genetic control of transcription in the human prefrontal cortex
- Carlo Colantuoni
- , Barbara K. Lipska
- & Joel E. Kleinman
-
Research Highlights |
 A way to save sickle cells
A way to save sickle cells
-
Article
| Open Access Multiple reference genomes and transcriptomes for Arabidopsis thaliana
Multiple reference genomes and transcriptomes for Arabidopsis thaliana
- Xiangchao Gan
- , Oliver Stegle
- & Richard Mott
-
Review Article |
 Central dogma at the single-molecule level in living cells
Central dogma at the single-molecule level in living cells
- Gene-Wei Li
- & X. Sunney Xie
-
Article |
 A conditional knockout resource for the genome-wide study of mouse gene function
A conditional knockout resource for the genome-wide study of mouse gene function
- William C. Skarnes
- , Barry Rosen
- & Allan Bradley
-
Letter |
 Transcriptomic analysis of autistic brain reveals convergent molecular pathology
Transcriptomic analysis of autistic brain reveals convergent molecular pathology
- Irina Voineagu
- , Xinchen Wang
- & Daniel H. Geschwind
-
Letter |
 A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity
A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity
Realizing the full potential of human embryonic stem cells (hESCs) in research and clinical applications requires a detailed understanding of the genetic network that governs their unique properties. A genome-wide RNA interference screen identifies a wealth of new regulators of self-renewal and pluripotency properties in hESCs. The transcription factor PRDM14, for example, is required for the maintenance of hESC identity and reprogramming of somatic cells to pluripotency.
- Na-Yu Chia
- , Yun-Shen Chan
- & Huck-Hui Ng
-
Letter |
 Pericytes regulate the blood–brain barrier
Pericytes regulate the blood–brain barrier
The blood–brain barrier (BBB) is made up of vascular endothelial cells and was thought to have formed postnatally from astrocytes. Two independent studies demonstrate that this barrier forms during embryogenesis, with pericyte/endothelial cell interactions being critical to regulate the BBB during development. A better understanding of the relationship among pericytes, neuroendothelial cells and astrocytes in BBB function will contribute to our understanding of BBB breakdown during central nervous system injury and disease.
- Annika Armulik
- , Guillem Genové
- & Christer Betsholtz
-
Article |
 MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake
MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake
The uptake of calcium by mitochondria has a central role in cell physiology, and an imbalance can trigger cell death. Now the first protein that is localized to the mitochondrion and is specifically required for calcium uptake has been identified. This protein, mitochondrial calcium uptake 1 (MICU1), represents the founding member of a set of proteins required for high-capacity calcium uptake. Its discovery should aid in the full molecular characterization of the mitochondrial calcium uptake pathways.
- Fabiana Perocchi
- , Vishal M. Gohil
- & Vamsi K. Mootha
-
News |
 Watching a gene at work
Watching a gene at work
How human proteins are made from DNA can be followed in real time.
- Brendan Borrell

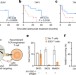 Identification of essential genes for cancer immunotherapy
Identification of essential genes for cancer immunotherapy Spinning threads
Spinning threads Presenting ENCODE
Presenting ENCODE