Abstract
Kawasaki disease (KD) is a systemic vasculitis and childhood febrile disease that can lead to cardiovascular complications. The diagnosis of KD depends on its clinical features, and thus it is sometimes difficult to make a definitive diagnosis. In order to identify diagnostic serum biomarkers for KD, we explored serum KD-related proteins, which differentially expressed during the acute and recovery phases of two patients by mass spectrometry (MS). We identified a total of 1,879 proteins by MS-based proteomic analysis. The levels of three of these proteins, namely lipopolysaccharide-binding protein (LBP), leucine-rich alpha-2-glycoprotein (LRG1), and angiotensinogen (AGT), were higher in acute phase patients. In contrast, the level of retinol-binding protein 4 (RBP4) was decreased. To confirm the usefulness of these proteins as biomarkers, we analyzed a total of 270 samples, including those collected from 55 patients with acute phase KD, by using western blot analysis and microarray enzyme-linked immunosorbent assays (ELISAs). Over the course of this experiment, we determined that the expression level of these proteins changes specifically in the acute phase of KD, rather than the recovery phase of KD or other febrile illness. Thus, LRG1 could be used as biomarkers to facilitate KD diagnosis based on clinical features.
Similar content being viewed by others
Introduction
Kawasaki disease (KD) is a systemic vasculitic disease whose etiology remains unknown. Moreover, KD can lead to cardiovascular complications such as coronary artery lesions (CALs)1,2. KD is the most frequent cause of childhood ischemic heart disease. The disease was first reported by Dr. T. Kawasaki, a Japanese pediatrician, in 19673, and took more than ten years to become widely known as a new disease entity. However, it is recognized today as the most frequent form of childhood vasculitis, apart from Henoch-Schönlein purpura4.
Symptoms of KD during the acute phase characteristically overlap in time, and disease manifestations are completed by the development of a fever refractory to antibiotic treatment, acute nonpurulent cervical lymphadenopathy, hyperemia of the bulbar conjunctiva, strawberry tongue, redness and cracking of the lips, hard edema of the hands and feet, and angiitis symptoms such as redness of finger tips by day 3 to day 7 after the onset. Moreover, in cases accompanied by development of CALs, echocardiography shows increased brightness of coronary arteries around day 10, and dilatation and aneurysm formation of the coronary arteries are detected between day 10 and day 14. Delaying definitive diagnosis of KD leads to development of CALs. However, there are no specific biomarkers for KD definitive diagnosis in clinical settings. The diagnosis of KD depends on its clinical features2. Therefore, it is often difficult to decide on the intervention for KD, especially in patients with incomplete clinical characteristics (incomplete KD), in the absence of a definitive diagnostic test5. Diagnostic methods based on biomarkers are necessary to assist physicians in diagnosing KD.
According to the 22nd national survey on KD in Japan6, there were more than 10,000 onset cases per year after 2005. KD cases have recently been increasing in number more rapidly. The morbidity is currently at 264.8 per 100,000 in the population of 0 to 4 year-olds in 2012. CALs used to develop in 25–30% of cases during the acute phase of KD, but since the introduction of intravenous immunoglobulin (IVIG) in the latter half of the 1980s, the onset rate of CALs has decreased to 9.3% at the acute phase, and 2.8% at the recovery phase6. The mortality was as high as 1% before 1974, but has decreased to 0.01% at present in Japan, probably because of intervention with IVIG7,8. Plasma exchange (PE) and anti tumor necrosis factor (TNF)-α therapy were also a safe and effective prophylactic measure against CALs in children with KD intractable to IVIG therapy9,10. Thus, safe and efficient therapeutics have been established for treating KD patients.
The pathogenesis of KD still remains obscure even almost 50 years after the disease’s discovery, although it may be summarized as abnormal activation of the immune system and panangiitis. However, the nature of KD therapeutics suggests that the serum may include a factor related to KD and CALs. Therefore, comprehensive analysis of the serum proteome of patients may be a key step in identifying candidate protein biomarkers that contribute to KD diagnosis and to the elucidation of KD pathogenesis. Serum samples are often used for biomarker discovery research. Previously, Zhang et al. performed two-dimensional electrophoresis (2-DE)-based analysis to compare the serum proteomes of normal children and KD patients by identifying differentially expressed proteins before and after IVIG therapy11. Although 2-DE is a fundamental technology for quantitative proteomics, it cannot characterize all of the protein components in serum. Recently, mass spectrometry (MS) has emerged as a powerful tool for discovering candidate protein biomarkers in proteomic studies, although cytokines and peptide hormones, which are very low-abundance proteins, are difficult to identify by MS.
In this study, we tried to comprehensively compare serum proteins from acute and recovery phase KD patients, using MS-based proteomic analysis for the identification of differentially expressed proteins. We subsequently validated KD-related proteins using specific antibodies. As a result, we identified four KD-related proteins, lipopolysaccharide-binding protein (LBP), leucine-rich alpha-2-glycoprotein (LRG1), angiotensinogen (AGT), and retinol-binding protein 4 (RBP4). These proteins are potentially useful for establishing a new diagnostic method to facilitate KD diagnosis based on clinical features.
Results
Identification of KD-related proteins using paired sera collected during the acute and recovery phases from two KD patients by MS-based proteomic analysis
MS-based proteomic analysis is a powerful tool for discovering disease related proteins in sera. In order to obtain accurate information related only to KD, we compared the proteomes of paired sera collected during the acute and recovery phases from two KD patients (No. 3 and No. 6). For this purpose, we performed label-free relative quantitation analysis with MS data as described in the Materials and Methods section (Fig. S1). The patients were male, aged 2 and 5 years, with typical KD; they exhibited five or more of the clinical features of KD and had lower than normal body temperatures due to IVIG treatment (Table S1). By using MS analysis, we identified and used 65,514 peptides derived from 1,813 proteins for relative quantitative analysis (Table S2). Among these, we found that 391 peptides derived from 56 proteins were upregulated and 102 peptides derived from 40 proteins were downregulated during the acute phase of KD compared to its recovery phase. In order to narrow down the KD-related proteins among these, we selected proteins that yielded at least five peptides that were either up- or downregulated during the acute phase of KD. Consequently, 20 proteins that were upregulated and six that were downregulated proteins in the acute phase compared to the recovery phase were selected as KD-related proteins (Table 1). Among them, ceruloplasmin (CP), plasminogen (PLG), alpha-1-antichymotrypsin (SERPINA3), and complement system proteins (Complement C1r subcomponent (C1R), Complement factor B (CFB), Complement component 9 (C9), Complement C1s subcomponent (C1S), Complement component 4-A (C4A), Complement component 4-B (C4B), and Complement component 7 (C7)) have been frequently detected in the acute phase of several diseases. We also detected serum amyloid A-1 protein (SAA1), which was previously reported to be KD-related12. Therefore, our MS-based proteomic analysis indicates precise differences in protein expression levels in the serum between the acute and recovery phases of KD. Previous work identified transthyretin (TTR) as a potential marker for molecular diagnosis and progression monitoring of KD11. However, we did not detect TTR in this study, likely because it was removed by the immune depletion column prior to MS analysis.
Validating candidate protein biomarkers for monitoring KD phases using western blotting
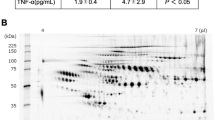
To experimentally refine potential serum biomarkers for use in KD diagnosis, we performed western blots to monitor the expression levels of some KD-related proteins in paired sera containing high-abundance proteins obtained from ten KD patients during the acute and recovery phases (Figs 1 and S2). We determined that the expression levels of three proteins (LBP, LRG1, and AGT) were significantly higher in acute phase serum samples than in recovery phase samples. In contrast, the expression level of RBP4 was significantly lower in acute phase serum samples than in recovery phase samples. These observations are consistent with the results of our MS-based proteomic analysis.
Western blot analysis of LBP, LRG1, AGT, and RBP4 were performed. The paired sera (collected separately during the acute and recovery phases) obtained from ten KD patients (Patient No. 1 to 10) were used in this experiment. The panels on the right show the expression level of protein in the serum of each patient determined with the western blots shown on the left. The horizontal lines and I bars indicate the means and SEMs, respectively. P values are used to compare expression between the acute and recovery phases of KD. Each mean ± SEM is as following; LBP: 4.2E + 07 ± 2.6E + 07 (acute) and 1.6E + 07 ± 1.0E + 07 (recovery) (p < 0.0020), LRG1: 6.6E + 07 ± 2.1E + 07 (acute) and 5.1E + 07 ± 2.6E + 07 (recovery) (p < 0.0059), AGT: 2.7E + 07 ± 4.6E + 06 (acute) and 1.8E + 07 ± 3.9E + 06 (recovery) (p < 0.0020), and RBP4: 2.6E + 07 ± 9.3E + 06 (acute) and 6.1E + 07 ± 2.4E + 07 (recovery) (p < 0.002). The gels/blots were cropped and the full-length gels and blots were included in Supplementary Fig. 5.
Validating potential protein biomarkers for monitoring KD phase using microarray enzyme-linked immunosorbent assays (ELISAs)
In order to carry out further experimental validation, we determined the expression levels of four proteins in the sera of KD patients and healthy subjects by microarray ELISAs, without depleting high-abundance proteins (Fig. 2A). Even when we examined a large number of patient sera, the concentrations of three proteins (LBP, LRG1, and AGT) were significantly higher in the acute phase than the recovery phase (p < 0.001), while the concentration of RBP4 was significantly lower in the acute phase compared to the recovery phase (p < 0.001) (Fig. 2, Table 2). In addition, receiver operating characteristic (ROC) curve analysis indicated that these KD-related proteins were useful for monitoring KD phase (Fig. S3). In particular, we observed that LBP and LRG1 expression levels in paired sera obtained during the acute and recovery phases from 42 KD patients decreased when their fever subsided (Fig. 3).
Panel A shows that expression levels of the four candidate biomarkers were significantly different in the acute phase of KD compared to the recovery phase of KD patients or healthy subjects. Panel B shows that the expression levels were significantly different in patients during the acute phase of KD compared to patients with other illnesses (G1: viral infections, G2: bacterial infections, and G3: autoimmune disease in childhood), except for LBP and AGT in G2. Microarray ELISAs were performed for LBP, LRG1, AGT, and RBP4, as described in the Materials and Methods section. The sera used in this experiment are described in Tables S1 and S3. The horizontal lines and I bars indicate means and the SEMs, respectively. P values are used for the comparison of the expression level among the acute and recovery phases of KD and other illnesses; ***p < 0.001, **p < 0.01, *p < 0.1, NS: non-significant. Each mean ± SD is described in Table 2.
Verification of candidate biomarkers for KD diagnosis
In order to evaluate the potential of the four KD-related proteins for use in distinguishing between KD and other childhood illnesses such as viral infections (G1), bacterial infections (G2), and autoimmune diseases (G3), we determined their expression levels in sera from patients suffering either from KD or these other illnesses using microarray ELISAs. The concentrations of three proteins (LBP, LRG1, and AGT) were significantly higher (p < 0.001) in the sera from the acute phase of KD than the other illnesses, except for LBP and AGT in G2. The concentration of RBP4 was significantly lower in the serum of acute phase of KD than the other illnesses (Fig. 2B). RBP4 was previously reported to increase after IVIG therapy in the serum of KD patients11. In this study, we found that the serum level of RBP4 was lower in acute phase than in recovery phase (two samples) of KD patients not subjected to IVIG therapy, or in samples from healthy subjects or those with other illnesses.
Morbidity of KD is high in 0–5 year-olds. Therefore, we evaluated the usefulness of these biomarkers in patients under 5 years of age. The conclusion of the ROC curve analysis was consistent with the results obtained in children aged from 0 to 5 years, comprising almost all typical KD patients (Fig. S4). Therefore, these results suggest that these proteins are potential serum biomarkers during the acute phase of KD. In addition, some diseases caused by adenovirus, Staphylococcus, and Streptococcus species infection mimic KD in clinical settings13. Therefore, we next evaluated whether these KD-related proteins were useful for distinguishing KD from these diseases. The concentrations of three proteins (LBP, LRG1, and RBP4) significantly changed in sera from the acute phase of KD, but not in the other diseases (Fig. 4). These results suggested that LRG1 should be a particularly effective serum biomarker for distinguishing KD from these illnesses.
Expression levels of the four candidate biomarkers were evaluated in the acute phase of KD and other infections that mimic KD, such as human adenovirus, Staphylococcus, and Streptococcus species. The serum concentrations of three proteins (LBP, LRG1, and RBP4) differed significantly in the acute phase of KD in comparison with these infections. The sera used in this experiment are described in Tables S1 and S3. The horizontal lines and error bars indicate means and SEMs, respectively. P values were used to evaluate the significance of differences between the acute phase of KD and these infections: ***p < 0.001; **p < 0.01; *p < 0.1; ns: non-significant.
Discussion
KD is characterized by acute systemic vasculitis in childhood. In clinical settings, early diagnosis of KD is crucial for preventing cardiovascular complications such as CALs. Because existing clinical criteria for KD are often insufficient for early and definitive diagnosis, biomarkers can help diagnosis of the disease and aid selection of the best treatment. Therefore, diagnosis using KD-related proteins will facilitate early and uniform diagnosis that is not reliant on the subjectivity and experience of physicians solely using clinical criteria.
Recently, many genetic studies of KD patients have been performed that found associations between genetic polymorphisms with susceptibility to KD and development of CALs2. Genes encoding inositol 1,4,5- three phosphorus acid 3-kinase C (ITPKC)14, caspase-3 (CASP3)15, B lymphocyte kinase (BLK), human leukocyte antigen (HLA), CD40 (CD40), and Fc fragment of Immunoglobulin G (IgG) low-affinity IIa receptor (FC-GR2A)16 were identified as products of KD susceptibility genes17. However, these genetic characteristics have not been studied sufficiently to understand their impact on KD pathogenesis and diagnosis. Therefore, it is difficult to diagnose patients with KD definitively by analyzing KD-susceptibility gene expression. In addition, no genetic biomarkers are available for promptly verifying a KD diagnosis in clinical settings. On the other hand, Kusuda et al. discovered KD-specific molecules that are linked to microbe-associated molecular patterns (MAMPs) derived from biofilms and that decrease in level following IVIG treatment, in the serum of KD patients18. It can be difficult, however, to distinguish between KD and bacterial infections solely by analyzing these molecules in clinical settings.
In contrast, serum biomarkers have the potential for wide clinical application through the development of rapid diagnostic techniques such as microarray ELISA. Some serum biomarkers are already used for clinical diagnosis of many diseases19. As for KD, blood vessel wall growth factor (VEGF)20,21 and N-terminal B-type natriuretic peptide (NT-proBNP) have previously been reported as candidate biomarkers17,22. However, these proteins still cannot be used as biomarkers for KD diagnosis in clinical settings. Clinical study results could not demonstrate adequate effectiveness.
MS-based proteomic analysis is often used for biomarker discovery. Although protein biomarkers are usually present at low concentrations in serum23, certain highly abundant serum proteins can obscure the presence of many other less abundant ones. A vast dynamic range of protein abundance, up to 12 orders of magnitude, complicates the identification of candidate biomarkers in the serum proteome using proteomics approaches23,24. Therefore, removing such highly abundant proteins is an essential first step when analyzing the serum proteome by liquid chromatography (LC)-MS/MS for biomarker discovery in order to reduce sample complexity25. Therefore, many prefractionation methods have been developed for biomarker discovery by shotgun LC-MS/MS26. In this study, we carried out prefractionation of the serum proteome using high performance liquid chromatography (HPLC) with a C4 reversed-phase column. We were able to achieve highly reproducible fractionation in this manner, and identified numerous peptides derived from many proteins at various concentrations in serum as being available for relative quantitation (Table S2). Consequently, we could search for KD-related candidates from among these numerous serum-derived peptides.
For effective biomarker discovery, it is also important to collect a good sample. Environmental factors and genetic backgrounds may affect the serum proteome, indicating that disease-specific factors are difficult to identify by comparison between different individuals. Therefore, KD-specific biomarkers for use in clinical settings have not been established in previous proteomic studies using many serum samples. To screen for KD-related proteins, in this study we obtained paired sera from two KD patients during the acute and recovery phases and subjected them to shotgun LC-MS/MS. Although the number of patients was small, we simply compared serum proteome from the same patient during both the acute and recovery phases, without needing to consider differences between two individuals. We could expect to obtain important information on KD-related proteins at the beginning of this study.
In this manner, we successfully obtained indications as to the identity of KD-related proteins for use in effective monitoring of KD phase. Based on these indications, we validated experimentally the differentially expressed proteins using several clinical samples and verified that these proteins are actually associated with KD. We thereby effectively demonstrated without wasting time, effort, or expense that, among these proteins, four in particular may be potential serum biomarkers for KD following experimental validation using microarray ELISAs with specific antibodies.
In particular, we determined that LBP and LRG1 are present at very high levels in serum from KD patients in the acute phase compared with both patients in the recovery phase and with healthy subjects. The blood levels of these proteins are high enough to detect precisely and quickly using microarray ELISAs. LBP and LRG1 may therefore be useful biomarkers for monitoring KD phase in clinical settings, although the availability of clinical samples is often limited. Additionally, RPB4 represents a promising potential biomarker for monitoring disease and therapeutic efficacy11.
Additionally, KD pathogenesis is probably mediated by various molecules. Therefore, an assay using multiple biomarkers with different specificities is more likely to yield a more definitive diagnosis than a single biomarker assay. Several potential serum biomarkers have already been developed for KD diagnosis17. Although these markers can reveal the intensity of inflammation in KD, it remains difficult to distinguish this condition from other diseases in clinical settings. By contrast, the expression pattern of each KD-related protein identified in this study may be of diagnostic value in distinguishing KD from other childhood diseases that mimic KD. Accordingly, these proteins should be validated within a prospective cohort including various childhood febrile diseases with similar or overlapping symptoms. Further large-scale studies will be necessary to confirm the diagnostic utility of these serum protein biomarkers and support this finding.
It is possible that these proteins may contribute to KD pathogenesis, although the relation between these proteins and KD remains to be elucidated. LBP plays an important role in the innate immune response to bacteria and fungi27. LBP transmits a signal within a cell through a receptor, CD14, on the cell membrane of phagocytic cells such as macrophages, and promotes the secretion of various inflammatory cytokines28. In fact, we observed high concentrations of LBP in serum derived from some patients with bacterial infections (G2) (Fig. 2B). The serum concentrations of inflammatory cytokines, such as TNF-α, interleukin 1, and interleukin 6, were remarkably high in KD patients compared to healthy subjects29,30. The pathogenesis of KD can be summarized as abnormal activation of the immune system. Therefore, this may indicate that bacterial infections followed by LBP upregulation contribute in part to the onset of KD. LRG1 is associated with fibrosis and blood vessel remodeling31. LRG1 has also been reported to be an inflammatory biomarker in autoimmune diseases32,33. Rheumatoid arthritis (RA) is an inflammatory disorder and an autoimmune disease, in which the body’s immune system mistakenly attacks the joints. LRG1 concentration was significantly elevated in the serum of RA patients compared to that of healthy subjects but decreased following anti-TNF therapy involving infliximab and etanercept32. Serum LRG1 concentration decreased significantly in all KD patients due to recovery in this study (Fig. 3), although anti-TNF therapy also has a beneficial effect on the systemic inflammatory process in KD34. These results suggest that the KD’s pathogenesis is abnormal activation of the immune system, like autoimmune disease. Therefore, these KD-related proteins may have potential use in elucidating KD pathogenesis, although further studies using cell models are needed.
Materials and Methods
Patients
We collected serum samples from patients admitted to the following hospitals in Japan: Yokohama City University Hospital and Medical Center, Kanagawa Children’s Medical Center, Showa General Hospital, Kobe Children’s Primary Emergency Medical Center, and Japanese Red Cross Wakayama Medical Center. Informed consent from either the patients or their guardians was obtained before blood samples were collected (Tables S1 and S3). In addition, the protocol of this survey and research plan has been approved by the Clinical Ethics Committee of Yokohama City University Medical Center (D1502021). This study was also performed with the approval of the Ethics Committee in each of the medical facilities. Informed consent was obtained from all patients and/or their guardians.
The subjects of this study were 64 patients with KD aged 0–12 years (55 serum samples were obtained in the acute phase and 51 in the recovery phase) (Table S1). KD diagnosis was performed according to clinical criteria2. The acute phase is defined as the febrile period before intervention. The recovery phase is defined as the afebrile period following intervention. 151 patients with other illnesses aged from 0 to 18 years, who were divided into group 1 (G1; viral infection), group 2 (G2; bacterial infection), and group 3 (G3; autoimmune disease), and thirteen healthy subjects aged 0–10 years were served as a control group (Table S3).
Identification of candidate protein biomarkers
In order to identify KD-related proteins, we performed shotgun LC-MS/MS followed by label-free relative quantitation of peptides in sera obtained from two patients with KD during the acute and recovery phases. A schematic overview of our MS-based proteomic analysis workflow is summarized in Fig. S1.
Sample preparation for MS analysis
We used a Human 14 Multiple Affinity Removal System (MARS) column (Agilent Technologies, Palo Alto, CA, USA) to remove 14 human proteins (albumin, IgG, antitrypsin, Immunoglobulin A (IgA), transferrin, haptoglobin, fibrinogen, alpha2-macroglobulin, alpha1-acid glycoprotein, IgM, apolipoprotein AI, apolipoprotein AII, complement C3, and transthyretin) that are highly abundant in serum. Each serum sample was diluted with buffer A (Agilent Technologies) for MARS column, and passed through a cellulose acetate spin filter (0.22 μm, Agilent Technologies). Employing a HPLC system, a Chromaster system (Hitachi, Tokyo Japan), the preparative serum sample was injected into a MARS column (10 × 100 mm), and the flow-through fractions were subsequently collected (Fig. S5). The proteins were concentrated by centrifugal ultrafiltration using a spin concentrator (5 K MWCO, 4 ml, Agilent Technologies) and the buffer was exchanged to 0.1% trifluoroacetic acid (TFA). Next, we carried out fractionation of the serum proteome using HPLC with a C4 reversed-phase column. After exchanging the buffer, each serum sample (400 μg) was injected into a Vydac C4 reversed phase column (4.6 × 250 mm, Grace Vydac, Columbia, MD, USA) on a Chromaster system. The HPLC separation conditions were as follows: solvent A, 0.1%(v/v) TFA; solvent B, 0.1%(v/v) TFA/100%(v/v) 2-propanol; flow-rate, 0.7 ml/min; gradient, 0–15 min (0% solvent B), 15–18 min (25% solvent B), 18–60 min (100% solvent B); detection wavelength, 280 nm; column oven temperature, 40 °C. Fractions were collected every 1 min from 24 to 44 min (20 fractions) after sample injection. Each fraction was divided into two equal aliquots that were lyophilized and stored at −80 °C until use. To prepare peptides for MS, each sample was dissolved in lysis buffer (2 M urea, 0.5 M thiourea, and 50 mM NH4HCO3), reduced with Tris (2-carboxyethyl) phosphine (TCEP) at a final concentration of 5 mM, and subsequently alkylated with methyl methanethiosulfonate (MMTS) at a final concentration of 20 mM. Protein solutions were subjected to proteolysis with bovine trypsin (final concentration, 15 ng/μl) (Promega, Madison, WI, USA) at 37 °C for 16 h. To stop hydrolysis, TFA was added to a final concentration of 1%(v/v), and precipitates were removed by centrifugation. Acidified hydrolysates were desalted using C18 StageTips with C18 Empore disks35 (3 M, St. Paul, MN, USA). Eluted peptides were completely lyophilized.
LC-MS/MS for label-free relative quantitation and identification of proteins
LC-MS/MS analysis was performed on a TripleTOF 5600 (AB Sciex, Foster City, CA, USA) coupled with a DiNa-AP (KYA Technologies, Tokyo, Japan). A pre-column HiQ sil C18W-3 (500 μm id × 1mm, KYA Technologies) and a nanoscale HiQ sil C18W-3 (100 μm id × 10 cm, KYA Technologies) were used. Peptide samples from each fraction were loaded on a reversed phase column and resolved at a flow rate of 200 nL/min with an acetonitrile/0.1% (v/v) formic acid gradient. Peptides were separated using a 145 min gradient program consisting of 5–40% buffer B for 120 min (buffer A: 2% acetonitrile, 98% water, 0.1% formic acid; buffer B: 80% acetonitrile, 20% water, 0.1% formic acid). Full-scan mass spectra were measured from m/z 400 to 1250 in positive-ion electro/spray ionization mode on a TripleTOF 5600 mass spectrometer operated in data-dependent mode. Label-free relative quantitation was performed using Progenesis LC-MS (v4.1, Nonlinear Dynamics, Durham, NC, USA). To identify peptides, peak lists were created using Progenesis LC-MS. The peak lists were used to search against human protein sequences (20,233 sequences) in the UniProt Knowledgebase (UniProtKB/Swiss-Prot) database (version Jan 2013) using MASCOT (v2.4.1, Matrix Science, London, UK). The search parameters were as follows: semitrypsin digestion with two missed cleavages permitted; variable modifications: N-terminal carbamylation, β-methylthiolate of cysteine, and oxidation of methionine; peptide charge of 2+, 3+, 4+; peptide mass tolerance for MS data ± 0.05 Da; and fragment mass tolerance ± 0.1 Da. We used a 1% overall false discovery rate (FDR) as a cutoff value to export our results from the database search using the software MASCOT. In addition, peptides that yielded a peptide ion score of greater than or equal to 30, were used for relative quantitation. Moreover, peptides that were either upregulated or downregulated during the acute phase of KD were extracted using the following parameters in Progenesis LC-MS: maximum fold change ≥2, analysis of variance (ANOVA) p < 0.001, and the average of normalized abundance (higher condition in Progenesis LC-MS) ≥500.
Experimental validation of candidate protein biomarkers
The expression levels of some KD-related proteins discovered by proteomic analysis in the serum were investigated using western blots and microarray ELISAs. For western blot analysis, the diluted sera collected from ten KD patients during the acute phase and the recovery phase (Patient No. 1 to 10) were separated on 12.5% polyacrylamide gels before being electroblotted on to polyvinylidene difluoride (PVDF) membranes. The membrane was incubated in Blocking One solution (Nacalai Tesque, Kyoto, Japan) to block nonspecific binding before being incubated in anti-LBP antibody (diluted 1:3000; GeneTex, San Antonio, TX, USA), anti-LRG1 antibody (diluted 1:5000; Abcam, Cambridge, UK), anti-AGT antibody (diluted 1:100, Immuno-Biological Laboratories, Gunma, Japan), and anti-RBP4 antibody (diluted 1:1000; Abcam). Primary antibody incubation was followed by incubation with horseradish peroxidase–conjugated secondary antibody (diluted 1:5000). Signals were visualized using the ECL western blot detection system (GE Healthcare UK, Buckinghamshire, UK). To detect labeled proteins, membranes were scanned on a LAS-4000mini imaging system (GE Healthcare, Little Chalfont, UK). Band intensities were quantitated using MultiGauge 3.0 software (Fujifilm, Tokyo, Japan).
For the microarray ELISAs, we analyzed a total of 270 serum samples. These samples were collected from 55 patients in the acute phase of KD, 51 in the recovery phase of KD, 151 with other illnesses, and 13 healthy subjects were analyzed (Tables 2, S1, and S3). In this study, the following ELISA kits were used: LRG1 ELISA kit (Immuno-Biological Laboratories), AGT ELISA kit (Immuno-Biological Laboratories), LBP ELISA kit (Hycult Biotech, Uden, Netherlands), and RBP4 ELISA kit (CycLex, Nagano, Japan). Sera were prepared and each assay procedure was performed according to each manufacturer’s instructions.
Statistical analysis
Statistical analysis was performed using GraphPad Prism (version 5.04, GraphPad Software, La Jolla, CA, USA). We used a Wilcoxon matched-pairs signed rank test for statistical analysis of our western blot results. As for our microarray ELISA results, we analyzed the serum protein levels that we measured using a Kruskal-Wallis test, followed by Dunn’s multiple comparison tests to determine statistical significance. We also generated ROC curves and area under the ROC curves (AUC) using GraphPad Prism. Experimental data derived are expressed as mean ± the standard error of the mean (SEM).
All methods were carried out in accordance with relevant guidelines and regulations. All experimental protocols were approved by the Clinical Ethics Committee of Yokohama City University Medical Center.
Additional Information
How to cite this article: Kimura, Y. et al. Identification of candidate diagnostic serum biomarkers for Kawasaki disease using proteomic analysis. Sci. Rep. 7, 43732; doi: 10.1038/srep43732 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Sundel, R. P. Kawasaki disease. Rheum Dis Clin North Am 41, 63–73, viii, doi: 10.1016/j.rdc.2014.09.010 (2015).
Kim, K. Y. & Kim, D. S. Recent Advances in Kawasaki Disease. Yonsei Med J 57, 15–21, doi: 10.3349/ymj.2016.57.1.15 (2016).
Kawasaki, T., Kosaki, F., Okawa, S., Shigematsu, I. & Yanagawa, H. A new infantile acute febrile mucocutaneous lymph node syndrome (MLNS) prevailing in Japan. Pediatrics 54, 271–276 (1974).
Barut, K., Sahin, S. & Kasapcopur, O. Pediatric vasculitis. Curr Opin Rheumatol 28, 29–38, doi: 10.1097/BOR.0000000000000236 (2016).
Sonobe, T. et al. Prevalence of coronary artery abnormality in incomplete Kawasaki disease. Pediatr Int 49, 421–426, doi: 10.1111/j.1442-200X.2007.02396.x (2007).
Kuwabara, M. et al. Cardiac lesions and initial laboratory data in Kawasaki disease: a nationwide survey in Japan. J Epidemiol 25, 189–193, doi: 10.2188/jea.JE20140128 (2015).
Newburger, J. W. et al. A single intravenous infusion of gamma globulin as compared with four infusions in the treatment of acute Kawasaki syndrome. N Engl J Med 324, 1633–1639, doi: 10.1056/NEJM199106063242305 (1991).
Patel, R. M. & Shulman, S. T. Kawasaki disease: a comprehensive review of treatment options. J Clin Pharm Ther 40, 620–625, doi: 10.1111/jcpt.12334 (2015).
Mori, M. et al. Efficacy of plasma exchange therapy for Kawasaki disease intractable to intravenous gamma-globulin. Mod Rheumatol 14, 43–47, doi: 10.1007/s10165-003-0264-3 (2004).
Sonoda, K., Mori, M., Hokosaki, T. & Yokota, S. Infliximab plus plasma exchange rescue therapy in Kawasaki disease. J Pediatr 164, 1128–1132 e1121, doi: 10.1016/j.jpeds.2014.01.020 (2014).
Zhang, L. et al. Monitoring of the serum proteome in Kawasaki disease patients before and after immunoglobulin therapy. Biochem Biophys Res Commun 447, 19–25, doi: 10.1016/j.bbrc.2014.03.108 (2014).
Mitani, Y. et al. Elevated levels of high-sensitivity C-reactive protein and serum amyloid-A late after Kawasaki disease: association between inflammation and late coronary sequelae in Kawasaki disease. Circulation 111, 38–43, doi: 10.1161/01.CIR.0000151311.38708.29 (2005).
Lloyd, A. J., Walker, C. & Wilkinso, M. Kawasaki disease: is it caused by an infectious agent? Br J Biomed Sci 58, 122–128 (2001).
Onouchi, Y. et al. ITPKC functional polymorphism associated with Kawasaki disease susceptibility and formation of coronary artery aneurysms. Nat Genet 40, 35–42, doi: 10.1038/ng.2007.59 (2008).
Onouchi, Y. et al. Common variants in CASP3 confer susceptibility to Kawasaki disease. Hum Mol Genet 19, 2898–2906, doi: 10.1093/hmg/ddq176 (2010).
Onouchi, Y. et al. A genome-wide association study identifies three new risk loci for Kawasaki disease. Nat Genet 44, 517–521, doi: 10.1038/ng.2220 (2012).
Parthasarathy, P., Agarwal, A., Chawla, K., Tofighi, T. & Mondal, T. K. Upcoming biomarkers for the diagnosis of Kawasaki disease: A review. Clin Biochem 48, 1188–1194, doi: 10.1016/j.clinbiochem.2015.02.013 (2015).
Kusuda, T. et al. Kawasaki disease-specific molecules in the sera are linked to microbe-associated molecular patterns in the biofilms. PLoS One 9, e113054, doi: 10.1371/journal.pone.0113054 (2014).
Stenman, U. H. Biomarker development, from bench to bedside. Crit Rev Clin Lab Sci 53, 69–86, doi: 10.3109/10408363.2015.1075468 (2016).
Maeno, N. et al. Increased serum levels of vascular endothelial growth factor in Kawasaki disease. Pediatr Res 44, 596–599, doi: 10.1203/00006450-199810000-00021 (1998).
Breunis, W. B. et al. Disruption of vascular homeostasis in patients with Kawasaki disease: involvement of vascular endothelial growth factor and angiopoietins. Arthritis Rheum 64, 306–315, doi: 10.1002/art.33316 (2012).
Cho, S. Y. et al. Adjuvant laboratory marker of Kawasaki disease; NT-pro-BNP or hs-CRP? Ann Clin Lab Sci 41, 360–363 (2011).
Anderson, N. L. & Anderson, N. G. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 1, 845–867 (2002).
Jacobs, J. M. et al. Utilizing human blood plasma for proteomic biomarker discovery. J Proteome Res 4, 1073–1085, doi: 10.1021/pr0500657 (2005).
Bellei, E. et al. High-abundance proteins depletion for serum proteomic analysis: concomitant removal of non-targeted proteins. Amino Acids 40, 145–156, doi: 10.1007/s00726-010-0628-x (2011).
Tessitore, A. et al. Serum biomarkers identification by mass spectrometry in high-mortality tumors. Int J Proteomics 2013, 125858, doi: 10.1155/2013/125858 (2013).
Blairon, L., Wittebole, X. & Laterre, P. F. Lipopolysaccharide-binding protein serum levels in patients with severe sepsis due to gram-positive and fungal infections. J Infect Dis 187, 287–291, doi: 10.1086/346046 (2003).
Myc, A. et al. The level of lipopolysaccharide-binding protein is significantly increased in plasma in patients with the systemic inflammatory response syndrome. Clin Diagn Lab Immunol 4, 113–116 (1997).
Takahashi, K., Oharaseki, T. & Yokouchi, Y. Pathogenesis of Kawasaki disease. Clin Exp Immunol 164 Suppl 1, 20–22, doi: 10.1111/j.1365-2249.2011.04361.x (2011).
Eleftheriou, D. et al. Management of Kawasaki disease. Arch Dis Child 99, 74–83, doi: 10.1136/archdischild-2012-302841 (2014).
Song, W. & Wang, X. The role of TGFbeta1 and LRG1 in cardiac remodelling and heart failure. Biophys Rev 7, 91–104, doi: 10.1007/s12551-014-0158-y (2015).
Serada, S. et al. iTRAQ-based proteomic identification of leucine-rich alpha-2 glycoprotein as a novel inflammatory biomarker in autoimmune diseases. Ann Rheum Dis 69, 770–774, doi: 10.1136/ard.2009.118919 (2010).
Fujimoto, M. et al. Leucine-rich alpha2 -glycoprotein as a potential biomarker for joint inflammation during anti-interleukin-6 biologic therapy in rheumatoid arthritis. Arthritis Rheumatol 67, 2056–2060, doi: 10.1002/art.39164 (2015).
Levin, M. & Burgner, D. Treatment of Kawasaki disease with anti-TNF antibodies. Lancet 383, 1700–1703, doi: 10.1016/S0140-6736(14)60131-8 (2014).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2, 1896–1906, doi: 10.1038/nprot.2007.261 (2007).
Acknowledgements
This work was supported by the Special Coordination Fund for Promoting Science and Technology, “Creation and Innovation Centers for Advanced Interdisciplinary Research Areas” from the Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Science and Technology Agency (JST). Aart from the submitted work, Masaaki Mori has received grants from Takeda Pharmaceutical Co., Ltd., Chugai Pharmaceutical Co., Ltd., Ono Pharmaceutical Co., Ltd., Mitsubishi Tanabe Pharma, AbbVie LLC, UCB Japan Co. Ltd., Astellas Pharma Inc., and Eisai Co., Ltd. Dr. Mori has also received lecture fees from MSD K.K and AbbVie LLC, and consulting fees from Daiichi Sankyo Co., Ltd. and Taisho Pharmaceutical Co., Ltd.
Author information
Authors and Affiliations
Contributions
Y.K., M.A. and A.O. performed MS-based proteomic analysis; Y.K., Y.I. and M.Matsuo performed biochemical studies; M.Y., T.K. and M.Mori performed statistical analysis; H.S., K.O., I.M., T.I. and S.Y. collected serum samples from patients; Y.K., M.Y., Y.I., M.A. and M.Mori analyzed data and wrote the manuscript. All authors discussed the results and commented on the manuscript. H.H. supervised the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Kimura, Y., Yanagimachi, M., Ino, Y. et al. Identification of candidate diagnostic serum biomarkers for Kawasaki disease using proteomic analysis. Sci Rep 7, 43732 (2017). https://doi.org/10.1038/srep43732
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep43732
This article is cited by
-
Serum levels of PDGF-CC as a potential biomarker for the diagnosis of Kawasaki disease
Italian Journal of Pediatrics (2024)
-
Characterization of serum protein expression profiles in the early sarcopenia older adults with low grip strength: a cross-sectional study
BMC Musculoskeletal Disorders (2022)
-
An atypical case of Kawasaki disease with severe pneumonia in a neonate
BMC Pediatrics (2022)
-
LRG1: an emerging player in disease pathogenesis
Journal of Biomedical Science (2022)
-
An Immunological Axis Involving Interleukin 1β and Leucine-Rich-α2-Glycoprotein Reflects Therapeutic Response of Children with Kawasaki Disease: Implications from the KAWAKINRA Trial
Journal of Clinical Immunology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.