Abstract
How does a type II inhibitor bind to/unbind from a kinase target is still a confusing question because the small molecule occupies both the ATP pocket and the allosteric pocket of the kinase binding site. Here, by using enhanced sampling simulations (umbrella sampling, US) and two-end-state free energy calculations (MM/GSBA), we systemically studied the dissociation processes of two distinct small molecules escaping from the binding pocket of p38 MAP kinase through the allosteric channel and the ATP channel. The results show that the unbinding pathways along the allosteric channel have much lower PMF depths than those along the ATP channel, suggesting that the allosteric channel is more favorable for the dissociations of the two inhibitors and thereby supporting the general understanding that the largest channel of a target is usually the entry/exit pathway for the binding/dissociation of small molecules. Interestingly, the MM/GBSA approach yielded similar PMF profiles compared with those based on US, a much time consuming approach, indicating that for a general study, such as detecting the important transition state of a ligand binding/unbinding process, MM/GBSA may be a feasible choice.
Similar content being viewed by others
Introduction
Human protein kinases regulate a variety of essential physiological processes, including proliferation, invasion, angiogenesis and metastasis, etc1,2,3,4, making them important targets for drug discovery. All protein kinases share a structurally conserved catalytic domain which is composed of two major sub-domains, namely the N-terminal and the C-terminal lobes5,6. The two lobes are connected through a flexible linker region (or hinge region). The activation loop belonging to the C-terminal lobe and adjacent to the linker region regulates the conformational transition between the “on” state (active conformation) and the “off” state (inactive conformation) of the kinases. The ATP-binding site is located in the cleft between the two lobes and the linker region. Most small molecule inhibitors of kinases are known as type I inhibitors which target the ATP-binding pocket in the active conformation. In recently years, the crystal structures of imatinib7, sorafenib8 and BIRB7969 have revealed another kind of kinase inhibitors that occupy both the ATP-binding pocket and the adjacent hydrophobic pocket (also called allosteric pocket) and thereafter were named as type II inhibitors10. The type II inhibitors can prevent the kinase activation by binding to the inactive conformations of kinases.
When a type I inhibitor occupies the ATP-binding pocket, the activation loop adopts the conformation that exposes the ATP-binding pocket completely. Then the entry/exit pathway of the type I inhibitor in the active kinase is defined as the ATP-pocket channel. Whereas, when a type II inhibitor targets an inactive kinase, the conformational transition of the activation loop and the conserved DFG (Asp-Phe-Gly) motif will shrink the ATP cleft and create an allosteric pocket. Thus, the ATP-pocket channel narrows and another entry/exit pathway named as the allosteric-pocket channel appears (Figure 1A). Numerous studies have focused on the ATP pocket for the dissociations of type I inhibitors11,12,13. For instance, Capelli et al. have studied two type I inhibitors unbinding from the ATP channel from VEGFR2 by using steered molecule dynamics (SMD) simulations12 and we have exploited the free energy profile change of a type I inhibitor targeting ALK tyrosine kinase against a series of drug resistant mutations along the ATP pathway by using adaptive biasing force (ABF) simulations13. However, which one is the real pathway for the type II inhibitors to move into/out of the target is still unclear.
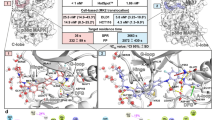
Superimposition of crystallized p38 MAP kinases complexed with ligands 1 (green stick and cartoon models, PDB code: 1KV1) and BIRB796 (pink stick and orange cartoon models, PDB code: 1KV2) (A).
The binding affinities of the ligands to p38 MAP kinase are very different as reported by reference [9], where the same part of the two molecules are colored in red (B). Surface models were shown to highlight (yellow region) the allosteric channels (panels C and E for 1 and BIRB796, respectively) and ATP channels (panels D and F for 1 and BIRB796, respectively).
p38 MAP kinase is one of the most investigated kinases targeted by type II inhibitors and the crystal structures in complex with type II inhibitors are also available (PDB entries: 1KV1 and 1KV2)9. Another reason why p38 MAP kinase can be used as a model system is that its allosteric pocket has been frequently studied14,15,16,17,18. For example, Wang et al. have characterized the free energy landscape of the allosteric pocket of p38 MAP kinase transferred from the DFG-in state to the DFG-out state by using normal mode analysis combined with umbrella sampling (NMA-US)18. Thereby, in this study, enhanced sampling simulations (umbrella sampling, US) were employed to investigate how two type II inhibitors (1 and BIRB796 shown in Figure 1) unbind from the active pocket through the ATP channel and the allosteric channel and then the Weighted Histogram Analysis Method (WHAM) was used to generate the potential of mean force (PMF) along the unbinding pathways. Meanwhile, the Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) method was also used to calculate the binding free profiles of the studied inhibitors along the unbinding pathways for a comparison.
Methods
Conventional Molecular Dynamics (MD) Simulations
The crystal structures of two type II inhibitors (1 and BIRB796) in complex with p38 MAP kinase (PDB entries: 1KV1 and 1KV29) were employed as the initial structures for the conventional MD simulations. The inhibitors were optimized and then the electrostatic potentials were calculated at HF/6-31G* level of theory supported in Gaussian 09. The partial charges were obtained by fitting the electrostatic potentials using the RESP technique19.
The AMBER03 force field20 was employed for the protein and the general AMBER force field (gaff)21 was used for the ligands. 7 Na+ were added to both the systems to neutralize the redundant net charges. Each system was immersed into a rectangular TIP3P water box22 that is extended 12 Å away from any solute atoms. The Particle Mesh Ewald (PME)23 algorithm was used to handle the long-range electrostatic interactions and the SHAKE24 algorithm was employed to constrain the covalent bonds involving hydrogen atoms. The time step was set to 2 fs. The cutoff of 10 Å was used for the short range interactions (van der Waals and electrostatic interactions).
Before MD simulations, steepest descent energy minimization (1000 cycles) and conjugate gradient energy minimization (4000 cycles) were carried out to optimize the systems. Then in the phase of MD simulations, each system was gradually heated from 0 to 300 K over the first 50 ps and followed by another 50 ps equilibrium simulation in the NPT ensemble (T = 300 K and P = 1 atm). In the two stages of MD simulations, the heavy atoms of the protein backbone were restrained with the elastic constant of 5 kcal/mol·Å2. Finally, a 10 ns production run without any constrain was performed in the NPT ensemble (T = 300 K and P = 1 atm). All the molecular mechanics (MM) minimizations and MD simulations were performed using the sander module in AMBER1125.
Umbrella Sampling Simulations
It is well known that the simulated systems are easily trapped in local minima and the sampling of some conformational transition processes, such as the unbinding process of a ligand, becomes a very hard task for conventional MD simulations. Thereby, it might need even millisecond level of conventional MD simulations to investigate the transition process for a small system26,27. Fortunately, the enhanced sampling methods, such as US28,29,30,31, metadynamics32,33 and adaptive biasing force (ABF)34,35, emerge as smart approaches to solve this problem, through adding either biasing potentials or biasing forces at the certain position of the reaction coordinate (RC) to enhance the sampling of the regions involved in high potential barriers. Take US as an example, to fully investigate the RC, the whole RC should be divided into a series of continuous windows. For convenience, harmonic potential, as shown in the equation below, is added to the original potential (unbiased potential) in each window to drive the system from one thermodynamic state to another.

where ui is the biased potential with respective to the current position r. ri is the reference position in window i and ki is the elastic constant used to pull the biased molecule out of the binding pocket. Here, an elastic constant of 5 kcal/mol·Å2 was used in all the US simulation windows. By adding the biasing potentials, some transition states with high energy barriers can also be fully sampled. To construct the potential of the mean force (PMF) along the RC, WHAM36,37 was employed to rebuild the biased probability distribution to a normal one. Herein, the RC was separated into 2000 bins for the WHAM calculation after each cycle of the US simulation (a 41 ns US run was defined as a cycle as shown below). The tolerance for iteration was set to 0.0001 to get convergent PMF curves. The temperature was set to 300 K to keep consistence with the simulation temperature.
In this study, two sets of US simulations were carried out to simulate the unbinding processes of two type II inhibitors through the ATP channel and the allosteric channel. During the simulations, the distance between one atom in receptor (O in Ile80 in 1KV1 for the allosteric pathway, Cα in Gln304 in 1KV1 for the ATP pathway, Cα in Gly81 in 1KV2 for the allosteric pathway and O in Arg66 in 1KV2 for the ATP pathway) and another atom in ligand (C8 in 1 in 1KV1 for the allosteric pathway, C1 in 1 in 1KV1 for the ATP pathway, N1 in BIRB796 in 1KV2 for the allosteric pathway and C4 in BIRB796 in 1KV2 for the ATP pathway) was selected as the RC and the vector between the two atoms represents the channel direction (the allosteric channel in Figures 1C and 1E and the ATP channel in Figures 1D and 1F). Each set of the simulations contains 41 simulation windows with 0.5 Å in length for each and the RCs were extended 20 Å away from the initial distances. For each window, 17 ns and 10 ns US simulations were preformed for the systems of 1 (1KV1) and BIRB796 (1KV2), respectively, to converge the sampling. Thereby, approximate 700 ns (697 ns) and 410 ns US simulations were preformed for systems 1 and BIRB796, respectively, which both contain the simulations for the ATP channel and allosteric channel and a total of >2.2 μs US simulations were performed in this study. To prevent the drifting of the systems, a restraint of 5 kcal/mol·Å2 was added to the heavy atoms that are 18 Å away from the ligands in the initial structures.
It has been proposed to use the constrained scheme for the calculations of absolute binding free energy38,39,40, but in this study we did not try to use these methods to calculate the binding free energy due to the fact that the constrained simulations may miss important metastable states when unbinding a ligand from the active site based on the prior determined straight RC13. Nevertheless, the purpose of this study is to reveal which pathway is more favorable for the ligand dissociation/entry and it is true that the difference of the absolute binding free energies is always reflected in the separation degree of freedom41. Therefore, the PMF calculated here for the comparison is actually the PMF depth rather than the absolute binding free energy.
MM/GBSA Binding Free Energy Calculation
To give a comparison with the probability-based PMF estimated from US simulations (estimated by WHAM), two-end-state free energy calculations were preformed directly based on the trajectories derived from the US simulations. Here, the widely used MM/GBSA methodology was employed for the two-end-state free energy calculation42,43,44,45,46,47,48,49,50. The binding free energy (ΔGbind) between a ligand and a receptor can be calculated according to the following equations47.



where ΔEMM is the gas-phase interaction between protein and ligand, including the electrostatic and the van der Waals energies; ΔGGB and ΔGSA are the polar and non-polar components of the desolvation free energy, respectively; -TΔS is the change of conformational entropy upon ligand binding at temperature T, which was not considered here due to the expensive computational cost and low prediction accuracy44. Here, the electrostatic solvation energy (ΔGGB) was calculated by the GB model developed by Onufriev (igb = 2)51, because this GB model usually outperforms the other GB models used in AMBER44. The exterior dielectric constant was set to 80 and the solute dielectric constant was set to 1. The non-polar contribution of the desolvation energy (ΔGSA) was estimated from the solvent accessible surface area (SASA) using the LCPO algorithm: ΔGSA = 0.0072 × ΔSASA + 0.0052.
Results and discussion
Convergence of the simulated systems
In order to equilibrate the systems, conventional MD simulations were performed for the complexes of p38 MAP kinase before the US simulations. As shown in Figure S1 in the supporting information, the low root mean square deviations (RMSDs, <1.5 Å on average) of the backbone heavy atoms with respect to the crystal structures suggest that the studied systems are stable throughout the 10 ns conventional MD simulations and thereby suitable for the following US analyses. The last snapshots of the conventional MD trajectories were chosen as the initial conformations for the US simulations.
To guarantee the sampling convergence of the US simulations, 17 ns and 10 ns US simulations for each window were preformed for the systems of 1 and BIRB796, respectively and the convergence of PMF was checked after each nanosecond of simulations. As shown in Figure 2, 17 and 10 curves were plotted for the systems of 1 (Figures 2A~2D) and BIRB796 (Figures 2E~2H), respectively. It can be found that the system of 1 converged after approximate 10 ns of the US simulations and 5 ns of the system BIRB796, with the PMF difference less than 1 kcal/mol (the PMF values were estimated based on 15 ~ 20 Å of the RCs). Interestingly, the PMF curves estimated by MM/GBSA showed convergence at the beginning of calculation. As shown in Figures 2C~2F, all the curves overlap in the same region and similar behaviors were found between the US and the MM/GBSA based PMF curves. For instance, when BIRB796 dissociates from the allosteric pathway, a low energy barrier was found in both the US results (Figure 2E) and the MM/GBSA results (Figure 2G) at ~5 Å of the RC, suggesting that MM/GBSA with much less computational cost is a feasible approach to calculate the free energies along a RC53. Nevertheless, due to the natural characteristics of the algorithm, the free energies calculated by MM/GBSA are much larger than those calculated by US (WHAM) and the experimental data (Table 1), most of which may be attributed to the ignorance of the contributions of the conformational entropies to the binding free energies54.
Binding free energies along allosteric and ATP channels
As shown in Figure 1, the same core structures (red fragments in panel B) and binding poses (panel A) were employed by the two inhibitors (1 and BIRB796). However, the extended fragments of BIRB796 (blue fragments) greatly enhances its binding affinity (from 1160 nM to 0.1 nM) to p38 MAP kinase as reported by Pargellis et al.9. As shown in Table 1 and Figure 3, the ranks of the binding free energies along both the allosteric pathway and the ATP pathway predicted by US (WHAM) and MM/GBSA are consistent with that of the experimental data. Compared with the US results, the free energies predicted by MM/GBSA along the two directions (allosteric and ATP pathways) are more consistent, with the difference of the binding free energies less than the standard deviations, but are much larger and unable to reach the flat phase of the PMF after the inhibitors dissociated from the target, especially the PMF curves of BIRB796 (Figure 3B cyan and red curves), which increase continuously after the molecule getting out of the binding pocket (Figures 5E and E′). As mentioned above, this phenomenon may be explained by the intrinsic limitations of MM/GBSA, which employs a parameterized implicit solvent model and uses some arbitrary parameters (post-processing parameters) to calculate the free energy, such as using different salt concentration and dielectric constants (such as εsolute = 1 and εsolvent = 80)54,55, implying that there is still interaction even the ligand moves far away from the receptor (20 Å). However, US is a probability based method and the probability distribution can be derived directly from the simulation trajectory which depends only on the original simulation settings (without using another post-processing method), for example, using the explicit solvent model (TIP3P water) which is closer to a real system. Therefore, the PMF curves derived from US should be closer to the reality and the experimental data (Table 1).
As shown in Table 1 and Figure 3A, the PMF values derived from the US simulations along the two channels are quite different for the two inhibitors (with the energy difference more than 6 kcal/mol and much larger than the standard deviations). Therefore, the allosteric channel is apparently more favorable for the dissociations of the two inhibitors. Interesting, along either pathway, the binding affinities of the two inhibitors can be correctly ranked, such as −19.03 kcal/mol for 1 versus −26.06 kcal/mol for BIRB796 along the ATP channel and −12.34 kcal/mol for 1 versus −19.57 kcal/mol for BIRB796 along the allosteric channel. That is to say, correct ranking for different molecules may be always resulted once a same dissociation pathway was employed, though the binding free energies along different pathways may be quite different. Nevertheless, the binding free energy difference (ΔPMFUS) between the two molecules (1 and BIRB796), which is 7.03 or 7.23 kcal/mol along the ATP or allosteric pathway, (Table S1) is in good consistence with the experimental binding affinity difference of 5.56 kcal/mol (ΔΔGexp), well supporting the opinion that same dissociation pathway should be employed when determining the relative binding affinities of a set of ligands.
Dissociation processes of 1 and BIRB796 along the allosteric and ATP channels
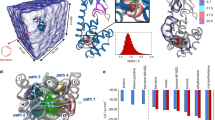
To determine the favorable pathway for the two inhibitors (1 and BIRB796) unbinding from/binding to the target, PMFs derived from the last 1 ns US simulations were connected between the allosteric pathway and the ATP pathway as shown in Figures 4G and 5F. The lowest value of the PMFs (0 kcal/mol) were calibrated to 0 Å on the RCs to denote the bound states of the inhibitors. The RCs were extended from 0 to −20 Å and 0 to 20 Å to represent the inhibitors 20 Å away from the ATP channel side and the allosteric channel side of the binding pocket, respectively. In theory, the PMF values should be equal everywhere once the ligands move far away from the binding site (20 Å away from the binding pocket) due to the isotropic bulk. However, as shown in Figures 4G and 5F, the PMF values along the ATP pathway are actually much higher than those along the allosteric pathway for the two ligands (both more than 6 kcal/mol as shown in Table 1), indicating that it may be more favorable for the small molecules dissociating along the allosteric pathway due to the much lower PMF depth (0 ~ 20 Å of the RC in Figures 4G and 5F) and more similar binding free energies to the experimental data (Table 1). As shown in Figures 1C~1F, the allosteric pocket (panels C and E) is obviously larger than the ATP pocket (panels D and F), explaining why much lower PMFs were produced when the inhibitors dissociated from the allosteric pathway and supporting the general opinion that the largest pocket pathway is usually considered as the binding/unbinding pathway13,41,56. The reason why the PMFs along the ATP pathway are much larger than those along the allosteric pathway may be partially related to the hard sampling of some associated metastable states of the simulations. As discussed by St-Pierre et al., it will be hard to generate reliable PMF curves when selecting an unfavorable pathway even the convergence of sampling is achieved57. Nevertheless, the hard sampling of certain pathways may be an efficient way to find the favorable pathway for the ligand entry/dissociation. However, as mentioned above, the PMFs derived from MM/GBSA are more consistent and thereby making it hard to determine which way is more favorable for the binding/unbinding of the inhibitors.
As shown in Figures 4G and 5F, different unbinding processes were observed for the two molecules though similar core structures were employed by them (red fragments in Figure 1B). When 1 escapes through the allosteric pathway, an obvious energy barrier is shown at point C of the PMF profile (5.2 Å of the RC in Figure 4G), where the methyl head (1 in Figure 1B) gradually gets out of the binding pocket (Figures 4B and 4C) to the more widely metastable outside binding site (6.3 Å of the RC in Figure 4E). As the large part of the molecule moves out of the binding pocket (Figure 4D), the ligand rapidly absorbs on the outside binding position (Figure 4E) with the PMF curve decreasing markedly (point E in Figure 4G). After unbinding from the outside binding position, 1 totally dissociates from the target (Figure 4F). Alternatively, no energy barrier is observed when 1 dissociates from the ATP pathway (0 ~ −20 Å of the RC in Figure 4G). It may be very unfavorable for the ligand unbinding through the ATP pathway because the binding channel is much longer (~13 Å of the ATP channel versus ~5 Å of the allosteric channel, Figure 4G) and narrower (Figures 1C and 1D) compared with the allosteric pathway. As shown in Figure 4G, three stages of steep upgrading of the PMF (regions of A′~B′, B′~C′ and D′~E′ of the RC) are observed before 1 dissociating from the ATP channel of the target, where the conformation of the ligand is pulled from the curved state to flat state through the three stages of dissociation (Figures 4A′~4F′).
Compared with the PMF profile of 1, the PMF curve of BIRB796 is much smoother with only a low energy barrier at approximate 5 Å of the RC (Figure 5B) when the ligand unbinds from the allosteric pathway, where the conformation of the ligand is pulled to be nearly flat which corresponds to a high strain energy of the molecule. However, a rotation phenomenon is observed at the next time step probably because of the high conformational energy of BIRB796. The ligand opens the P-loop region of the kinase (blue region in Figure 5C) to relax the high strain energy of the flatted ligand which corresponds to a local minimum of the system as shown in C point of the PMF curve in Figure 5F. After that, the molecule dissociates vertically to the P-loop region to the bulk (Figures 5C and 5D). As for the ATP channel, a continued rising PMF without any plain state is observed when the molecule dissociates from the ATP channel (Figure 5F). Although the rising rate of the PMF (slope) seems to be moderated (point C′ of Figure 5F) when the N and O containing heterocyclic tail (Figure 1B) gets out of the ATP channel (Figure 5C′), the PMF curve rises continuously till the bifurcated head, which contains a benzene and a methyl with the angle >120° connected to the double N hetero-ring (pink stick model in Figure 1A), leaves the narrow channel (after the D′ point of the PMF profile as shown in Figure 5F). Thereby, the large bifurcated head of BIRB796 is not favorable to dissociate from the ATP channel, though similar unbinding lengths of the allosteric (> 8 Å) and the ATP (~10 Å) channels are observed in the PMF profile in Figure 5F.
Conclusion
By using enhanced sampling simulations and two-end-state calculations, we systemically investigated the unbinding processes of two type II inhibitors escaping from p38 MAP kinase through two pathways, namely the ATP pathway and the allosteric pathway. Consistent with the general idea that the largest channel is usually the probably entry/exit pathway for a ligand binding/dissociation, we found that the PMF depths of the allosteric channel for the two inhibitors are both much lower than those of the ATP pathway, indicating that the inhibitors may dissociate/bind through the allosteric channel. Besides, although the ATP channel may be not a favorable pathway for the ligands dissociation, right ranking of the binding free energies of the two distinct inhibitors was also observed (PMFs based on US), implying that similar (binding/unbinding) pathway should be used for the binding affinity comparison.
Moreover, although MM/GBSA may result in much larger binding free energies compared with the experimental data, it is much easier to gain convergent PMFs compared with the much time consuming US simulations and keeps similar shape between the PMF profiles based on US as well. Thereby, to detect important transition state, MM/GBSA may also be a feasible choice to calculate the binding free energies along a RC.
References
Cohen, P. Protein kinases-the major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 1, 309–315 (2002).
Manning, G. et al. The Protein Kinase Complement of the Human Genome. Science 298, 1912–1934 (2002).
Dancey, J. & Sausville, E. A. Issues and progress with protein kinase inhibitors for cancer treatment. Nat. Rev. Drug Discov. 2, 296–313 (2003).
Ripphausen, P. et al. Quo Vadis, Virtual Screening? A Comprehensive Survey of Prospective Applications. J. Med. Chem. 53, 8461–8467 (2010).
Huse, M. & Kuriyan, J. The conformational plasticity of protein kinases. Cell 109, 275–282 (2002).
Scapin, G. Structural biology in drug design: selective protein kinase inhibitors. Drug Discov. Today 7, 601–611 (2002).
Schindler, T. et al. Structural mechanism for STI-571 inhibition of Abelson tyrosine kinase. Science 289, 1938–1942 (2000).
Wan, P. T. C. et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116, 855–867 (2004).
Pargellis, C. et al. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat. Struct. Biol. 9, 268–272 (2002).
Liu, Y. & Gray, N. S. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2, 358–364 (2006).
Yang, L. J. et al. Steered molecular dynamics simulations reveal the likelier dissociation pathway of imatinib from its targeting kinases c-Kit and Abl. PloS One 4, e8470 (2009).
Capelli, A. M. & Costantino, G. Unbinding pathways of VEGFR2 inhibitors revealed by steered molecular dynamics. J. Chem. Inf. Model. (2014).
Sun, H. et al. Insight into Crizotinib Resistance Mechanisms Caused by Three Mutations in ALK Tyrosine Kinase using Free Energy Calculation Approaches. J. Chem. Inf. Model. 53, 2376–2389 (2013).
Frembgen-Kesner, T. & Elcock, A. H. Computational sampling of a cryptic drug binding site in a protein receptor: Explicit solvent molecular dynamics and inhibitor docking to p38 MAP kinase. J. Mol. Biol. 359, 202–214 (2006).
Filomia, F. et al. Insights into MAPK p38 alpha DFG flip mechanism by accelerated molecular dynamics. Bioorg. Med. Chem. Lett. 18, 6805–6812 (2010).
Badrinarayan, P. & Sastry, G. N. Sequence, Structure and Active Site Analyses of p38 MAP Kinase: Exploiting DFG-out Conformation as a Strategy to Design New Type II Leads. J. Chem. Inf. Model. 51, 115–129 (2011).
Yang, Y. et al. Molecular Dynamics Simulation and Free Energy Calculation Studies of the Binding Mechanism of Allosteric Inhibitors with p38 alpha MAP Kinase. J. Chem. Inf. Model. 51, 3235–3246 (2011).
Wang, J. et al. Exploring Transition Pathway and Free-Energy Profile of Large-Scale Protein Conformational Change by Combining Normal Mode Analysis and Umbrella Sampling Molecular Dynamics. J. Phys. Chem. B. 118, 134–143 (2013).
Bayly, C. I. et al. A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges-the Resp Model. J. Comput. Phys. 97, 10269–10280 (1993).
Duan, Y. et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24, 1999–2012 (2003).
Wang, J. M. et al. Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004).
Jorgensen, W. L. et al. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Darden, T. et al. Particle Mesh Ewald-an N.Log(N) Method for Ewald Sums in Large Systems. J. Comput. Phys. 98, 10089–10092 (1993).
Ryckaert, J. P. et al. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints-Molecular-Dynamics of N-Alkanes. J. Comput. Phys. 23, 327–341 (1977).
Case, D. A. et al. The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005).
Dror, R. O. et al. Activation Mechanism of the β2-Adrenergic Receptor. Proc. Natl. Acad. Sci. USA 108, 18684–18689 (2011).
Nygaard, R. et al. The Dynamic Process of β2-Adrenergic Receptor Activation. Cell 152, 532–542 (2013).
Torrie, G. M. & Valleau, J. P. Monte-Carlo Study of a Phase-Separating Liquid-Mixture by Umbrella Sampling. J. Comput. Phys. 66, 1402–1408 (1977).
Torrie, G. M. & Valleau, J. P. Non-Physical Sampling Distributions in Monte-Carlo Free-Energy Estimation-Umbrella Sampling. J. Comput. Phys. 23, 187–199 (1977).
Adcock, S. A. & McCammon, J. A. Molecular dynamics: Survey of methods for simulating the activity of proteins. Chem. Rev. 106, 1589–1615 (2006).
Kastner, J. Umbrella sampling. Wiley Interdiscip. Rev. Comput. Mol. Sci. 1, 932–942 (2011).
Laio, A. & Parrinello, M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA 99, 12562–12566 (2002).
Laio, A. & Gervasio, F. L. Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep. Prog. Phys. 71, 126601 (2008).
Darve, E. et al. Adaptive biasing force method for scalar and vector free energy calculations. J. Chem. Phys. 128, 144120–144132 (2008).
Hénin, J. et al. Exploring multidimensional free energy landscapes using time-dependent biases on collective variables. J. Chem. Theory Comput. 6, 35–47 (2009).
Souaille, M. & Roux, B. Extension to the weighted histogram analysis method: combining umbrella sampling with free energy calculations. Comput. Phys. Commun. 135, 40–57 (2001).
Kumar, S. et al. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 13, 1011–1021 (1992).
Woo, H.-J. & Roux, B. Calculation of absolute protein–ligand binding free energy from computer simulations. Proc. Natl. Acad. Sci. USA 102, 6825–6830 (2005).
Gumbart, J. C. et al. Standard Binding Free Energies from Computer Simulations: What Is the Best Strategy? J. Chem. Theory Comput. 9, 794–802 (2012).
Doudou, S. et al. Standard free energy of binding from a one-dimensional potential of mean force. J. Chem. Theory. Comput. 5, 909–918 (2009).
Sun, H. et al. P-loop Conformation Governed Crizotinib Resistance in G2032R-mutated ROS1 Tyrosine Kinase: Clues from Free Energy Landscape. PLoS Comput. Biol. 10, e1003729 (2014).
Sun, H.-Y. et al. Finding chemical drugs for genetic diseases. Drug Discov. Today 19, 1836–1840 (2014).
Sun, H.-Y. et al. Structural and energetic analyses of SNPs in drug targets and implications for drug therapy. J. Chem. Inf. Model. 53, 3343–3351 (2013).
Hou, T. J. et al. Assessing the Performance of the MM/PBSA and MM/GBSA Methods. 1. The Accuracy of Binding Free Energy Calculations Based on Molecular Dynamics Simulations. J. Chem. Inf. Model. 51, 69–82 (2011).
Hou, T. J. & Yu, R. Molecular dynamics and free energy studies on the wild-type and double mutant HIV-1 protease complexed with amprenavir and two amprenavir-related inhibitors: Mechanism for binding and drug resistance. J. Med. Chem. 50, 1177–1188 (2007).
Huo, S. H. et al. Molecular dynamics and free energy analyses of cathepsin D-inhibitor interactions: Insight into structure-based ligand design. J. Med. Chem. 45, 1412–1419 (2002).
Kollman, P. A. et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 33, 889–897 (2000).
Kuhn, B. & Kollman, P. A. Binding of a diverse set of ligands to avidin and streptavidin: an accurate quantitative prediction of their relative affinities by a combination of molecular mechanics and continuum solvent models. J. Med. Chem. 43, 3786–3791 (2000).
Wang, J. M. et al. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J. Am. Chem. Soc. 123, 5221–5230 (2001).
Xue, W. W. et al. Molecular modeling study on the resistance mechanism of HCV NS3/4A serine protease mutants R155K, A156V and D168A to TMC435. Antiviral Res. 93, 126–137 (2012).
Onufriev, A. et al. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins: Struct., Funct., Bioinf. 55, 383–394 (2004).
Weiser, J. et al. Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO). J. Comput. Chem. 20, 217–230 (1999).
Bai, F. et al. Free energy landscape for the binding process of Huperzine A to acetylcholinesterase. Proc. Natl. Acad. Sci. USA 110, 4273–4278 (2013).
Sun, H. et al. Assessing the Performance of MM/PBSA and MM/GBSA Methods. 4. Accuracies of MM/PBSA and MM/GBSA Methodologies Evaluated by Various Simulation Protocols using PDBbind Data Set. Phys. Chem. Chem. Phys. 16, 16719–16729 (2014).
Sun, H. et al. Assessing the performance of MM/PBSA and MM/GBSA methods. 5. Improved docking performance using high solute dielectric constant MM/GBSA and MM/PBSA rescoring. Phys. Chem. Chem. Phys. 16, 22035–22045 (2014).
Pan, P. et al. Molecular Principle of Topotecan Resistance by Topoisomerase I Mutations through Molecular Modeling Approaches. J. Chem. Inf. Model. 53, 997–1006 (2013).
St-Pierre, J. F. et al. Use of Umbrella Sampling to Calculate the Entrance/Exit Pathway for Z-Pro-Prolinal Inhibitor in Prolyl Oligopeptidase. J. Chem. Theory Comput. 7, 1583–1594 (2011).
Acknowledgements
This study was supported by the National Science Foundation of China (21173156), the National Basic Research Program of China (973 program, 2012CB932600), the Research Fund for the Doctoral Program of Higher Education of China (20123201110017), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), Jiangsu Key Laboratory for Carbon-Based Functional Materials and Devices and Collaborative Innovation Center of Suzhou Nano Science and Technology.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: T.H. Performed the experiments: H.S., S.T., S.Z., Y.L., D.L., L.X., M.S. and P.P. Analyzed the data: H.S., S.T., S.Z., Y.L., D.L., L.X., M.S. and P.P. Contributed reagents/materials/analysis tools: H.S., S.T., S.Z., Y.L., D.L., L.X., M.S. and P.P. Wrote the paper: H.S. and T.H.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Suppporting Materials
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Sun, H., Tian, S., Zhou, S. et al. Revealing the favorable dissociation pathway of type II kinase inhibitors via enhanced sampling simulations and two-end-state calculations. Sci Rep 5, 8457 (2015). https://doi.org/10.1038/srep08457
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep08457
This article is cited by
-
Oncogenic G12D mutation alters local conformations and dynamics of K-Ras
Scientific Reports (2019)
-
Molecular Modeling of ALK L1198F and/or G1202R Mutations to Determine Differential Crizotinib Sensitivity
Scientific Reports (2019)
-
Structure-Based Virtual Screening of High-Affinity ATP-Competitive Inhibitors Against Human Lemur Tyrosine Kinase-3 (LMTK3) Domain: A Novel Therapeutic Target for Breast Cancer
Interdisciplinary Sciences: Computational Life Sciences (2019)
-
A molecular dynamics investigation of CDK8/CycC and ligand binding: conformational flexibility and implication in drug discovery
Journal of Computer-Aided Molecular Design (2018)
-
How Does the L884P Mutation Confer Resistance to Type-II Inhibitors of JAK2 Kinase: A Comprehensive Molecular Modeling Study
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.