Abstract
The villous trees of human placentas delineate the fetomaternal border and are complex three-dimensional (3D) structures. Thus far, they have primarily been analyzed as thin, two-dimensional (2D) histological sections. However, 2D sections cannot provide access to key aspects such as branching nodes and branch order. Using samples taken from 50 normal human placentas at birth, in the present study we show that analysis procedures for 3D reconstruction of neuronal dendritic trees can also be used for analyzing trees of human placentas. Nodes and their branches (e.g., branching hierarchy, branching angles, diameters and lengths of branches) can be efficiently measured in whole-mount preparations of isolated villous trees using high-end light microscopy. Such data differ qualitatively from the data obtainable from histological sections and go substantially beyond the morphological horizon of such histological data. Unexpectedly, branching angles of terminal branches of villous trees varied inversely with the fetoplacental weight ratio, a widely used clinical parameter. Since branching angles have never before been determined in the human placenta, this result requires further detailed studies in order to fully understand its impact.
Similar content being viewed by others
Introduction
The villous tree is the core part of the functionally relevant microarchitecture of the human placenta1,2,3,4. The surface of the villous tree is covered by the villous trophoblast, a syncytialized epithelium in direct apical contact with maternal blood that controls fetomaternal exchange.
Tree-like structures are generally characterized by nodes, which are cornerstones of a complex three-dimensional (3D) pattern of branches5. Due to the 3D nature of their branching structure, advanced morphological analysis of trees requires 3D analysis. The correlation of function with 3D structure of trees has been convincingly demonstrated in the field of neuroscience. The digital variants of camera lucida techniques have become the gold standard of computer-assisted quantitative morphological and functional 3D reconstruction of neuronal dendritic trees6,7,8,9. Completely new research fields were e.g. opened by the recent finding of the group of TC Südhof that neuroligin-3 mutations in autism are associated with altered 3D dendritic trees and thus altered neuronal connectivity10. Conceptional understanding of neuronal diversity and connectivity was revolutionized by this type of 3D analysis of dendritic trees11. Cortical neurons in the brain are classified by the 3D structure of their dendritic trees12,13. Moreover, such 3D morphological and multiple reconstruction of cortical neuronal axons and dendritic trees are an important tool to reconstruct the connectome of whole cortical columns, one of the most vivid fields of current neuroscience14. Similarly, such 3D analysis of the structure of placental villous trees should provide an important link between the mechanisms of villous branching4,15 and the capability of the placenta to adapt to varying functional requirements during pregnancy.
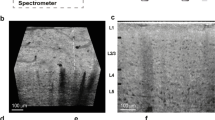
The quantitative histological analysis of the villous trees of thuman placentas has usually been based on thin two-dimensional (2D) histological sections that must be statistically representative 2D samples of the 3D space under examination, allowing mathematically founded statements on elements of the 3D space by the analysis of thin 2D histological sections16,17. However, all analyses of thin 2D histological sections of the human placenta suffer from the fact that nodes and, thus, the branching structure itself are excluded from the analysis. Nodes are not visible on thin 2D histological sections; there is not even a histological nomenclature for nodes. This is exemplified in Figure 1 by a comparison of the histology of a thin 2D histological section with the 3D aspect of an isolated villous tree of a human placenta.
Comparison of the histology of villous trees of the human placenta and their 3D aspects.
(a,b) Comparison of the microscopic aspects of a thin (4–6 μm) histological section of a human placenta after staining with hematoxylin/eosin (a) with the microscopic aspects of a whole-mount isolated villous tree after staining with hematoxylin (b). The scale bars in a and b are 250 μm. (a) Various cross- and longitudinal sections of villi can be recognized. The stromal architecture inside the sectioned villi is visible. The cross-sections of branches belong to an unknown number of villous trees. (b) A single villous tree is visible and branches are not sectioned. The hierarchical positions of nodes (branching points) and the branching topology can be recognized.
This disadvantage has not been changed by other morphological approaches, particularly by scanning electron microscopy or confocal microscopy. Scanning electron microscopy delivers a 3D impression. Because this impression is not easily quantified in x,y and z, scanning electron microscopy was primarily used to illustrate qualitative aspects of the villous tree. The latter was frequently combined with histology18 or with transmission electron microscopy4,19. Confocal microscopy can replace physical sectioning by optical sectioning; although software extensions allow post-microscopic 3D reconstruction20, confocal microscopy is primarily a technology that uses extremely thin, optical 2D histological sections. Three-dimensional reconstructions of capillary networks inside of only the last branch of the villous tree (terminal villi) of the human placenta were evaluated by confocal microscopy21,22,23. Nevertheless, the efficient 3D analysis of villous trees of human placentas across two or more generations of entire branches remains a challenge.
We have hypothesized that the computer-assisted modern camera lucida strategies known from neuroscience can be adapted to the analysis of isolated villous trees of human placentas. Furthermore, we have hypothesized that this adapted camera lucida approach will deliver quantitative 3D data of qualitatively novel information regarding the villous tree of the human placenta. To verify these hypotheses, we isolated villous trees from 50 normal human placentas by careful and diligent dissection under a dissecting microscope. Tissue samples were analyzed with a software, Neurolucida (MBF Bioscience, Williston, VT, USA), which was originally developed for the analysis of dendritic trees of neurons7,8,9 using settings adapted to the requirements of villous branching. In parallel, thin 2D histological sections (2D sections) of systematically and randomly sampled tissue blocks of a comparable collection of 50 normal human placentas were used to determine volume densities and absolute volumes of placental tissue components using stereology software, Stereo Investigator (MBF Bioscience).
We found that the villous tree of the human placenta can be analyzed with the Neurolucida approach, that novel and, thus far, unreachable variables can be extracted and that some of these novel data correlate with the fetoplacental weight ratio, which is a widely used clinical parameter.
Results
Three-dimensional analysis
The whole-mount preparations of villous trees of human placentas prepared for the present study could easily be viewed under a microscope in three dimensions (Fig. 2) and 3D camera lucida datasets could be established in parallel (Fig. 3); details of these procedures are outlined in the methods section. The 3D data obtained in the present study can be viewed and analyzed in various formats (Fig. 4). To distinguish the novel 3D approach of the present study from histological nomenclature, the parts of villous trees originating at the branching points (nodes) were called branches (b), not villi. Branches were classified by their distance to the nearest terminal end (terminal distance ordering of branches (bT)). The distance to the nearest terminal end was measured in nodes. Thus bT0 designates a branch in terminal position, bT1 a branch one node apart from the nearest terminal end, and, generally, bTX designated a branch X nodes apart from the nearest terminal end (Fig. 4).
Whole-mount preparations of villous trees of the human placenta preserve the three-dimensional (3D) character.
(a–o) Example of the 3D character (depth) of a whole-mount preparation of an isolated villous tree of a human placenta that was stained with hematoxylin. The example starts with an overview at low magnification ((a), 5× objective; scale bar = 250 μm) and shows a screenshot of a single microscopic field of view at working magnification ((b), 20× objective; scale bar = 25 μm). The red box in a and b highlights the approximative area of the field of view, which is shown in (c–o) (scale bar = 25 μm). The analysis used live microscopic images at working magnification, whereas the calibrated x, y, z-stage of the used microscope recorded the positions of the focus planes in x, y and z. (c–o) show an exemplary shift of the focus plane in the z-direction; screenshots were taken from the continuous stream every 10 μm (from 0 μm (c) to 120 μm(o)). Levels of z are given relative to the highest focus plane ((c); 0 μm) in the lower left corner of (c–o).
Computer-assisted viewing generates a quantitative digital three-dimensional (3D) replica of villous trees of the human placenta.
(a–y) Example illustrating the procedure by which the Neurolucida software assisted in generating a digital and quantitative 3D replica of a whole-mount preparation of an isolated villous tree of a human placenta. The entire instrument and software setup was calibrated such that the software recorded 3D-coordinates of all positions and mouse functions. Measurement and 3D-reconstruction were simultaneous processes. (a) shows an overview at low magnification (2× objective). The red box in a delineates the region of interest. The region of interest is larger than a single microscopic field-of-view at working magnification (20× objective). (b–y) Screenshots taken at working magnification (20× objective; scale bar = 25 μm on the top of each tile) while the tracing of the villous tree in the region of interest was in progress. The coordinates of the centerpoint of the yellow circle and the diameter of this circle were recorded while the villous tree was traced in x,y and z. The work flow started with the most proximal point of the region of interest (root point of the model, (b)). The yellow circle shown could be adjusted to the diameter of the villus in focus using the mouse wheel function. Its moving center defined the center line (yellow line) of each branch. Branching points (nodes; shown as yellow dots with associated red arrows) could be placed by clicking functions. Similarly, terminal ends were defined using the mouse wheel function and are shown as green dots with associated black arrows.
The 3D data can be viewed and analyzed in various formats.
(a–e) Example illustrating different graphing options of a three-dimensional data set obtained with Neurolucida software (MBF Bioscience) assisted reconstruction of a whole-mount preparation of a villous tree of a human placenta. (a) Overview of a hematoxylin-stained peripheral villous tree as viewed at low magnification (5× objective; scale bar = 250 μm). (b) Detailed view of a subtree within a single microscopic field of view at the magnification used for tracing the villous tree with Neurolucida software (20× objective; scale bar = 250 μm). The part of the visual field containing the subtree shown in (b) is labeled by rectangular red boxes in (a) and (c–e). (c–e) Various options to visualize the 3D data sets obtained by Neurolucida software. The center lines of traced branches are shown in the skeletonized view (c); 3D reconstructions of the traced branches based on diameters are shown in (d). The dendrogram view in (e) shows the total segmental length and hierarchical relations of branches. (f) Sketch of the working process in the Neurolucida software showing the tracing direction (from root to terminal ends) and the classification of branches by the terminal distance (from terminal ends to root, in numbers), the labeling of nodes/branching points and the determination of tree measures. (f) Was modified from the Neurolucida Explorer software with permission from MBF Bioscience.
We found none of the analyzed branches more than three nodes apart from the nearest terminal end. Accordingly, all analyzed branches were bT0, bT1, bT2 or bT3. The branching of the villous trees was primarily dichotomous. Trichotomous nodes rarely occured (we recorded 11 trichotomous nodes of 454 analyzed nodes). The villous trees under investigation could be fully reconstructed in 3D (Supplementary Movies S1–4).
There were statistically significantly (p < 0.001) more branches in positions bT0 and bT1 than in position bT2 (Fig. 5A). The dichotomous branching pattern of the villous tree was also indicated by a statistically significant (p < 0.05) linear correlation between the numbers of bT0 and bT1 branches of the villous trees (Fig. 5B). The mean ratio of all numbers of bT0 and bT1 branches across all peripheral villous trees analyzed in the present study was 1.30 (Fig. 5B; see also branching scheme in Fig. 4). In contrast, the ratio of the number of bT1 and bT2 branches varied over a wide range (Fig. 5C). The dichotomous branching pattern did not change across the positional transition from bT1 to bT2.
Branch numbers reveal construction rules of the peripheral villous tree of the human placenta.
(a–c) Number of branches. Each dot represents the number of branches of a single isolated villous tree fragment. (a) Number of branches (n[count]) stratified by the terminal distance to which the branches belong (bT0, bT1 and bT2). The mean, standard deviation and number of samples are shown. The number of branches in positions bT1 and bT2 was found to be statistically significantly lower than the number of branches in position bT0. The values of p and the number (n) of villous trees evaluated are given inside the graph. (b,c) Correlation of the number of branches in two neighboring terminal distance positions. The regression lines (straight line) are shown together with their 95% confidence interval (dashed lines). The p-value, expressing the statistically significant deviation of the slope from zero, R2 a measure of the goodness of fit and the number of villous trees evaluated (n) are given inside the graphs. (b) Number of bT1 (bT1_n) branches as a function of the number of bT0 (bT1_0) branches. (c) Number of bT2 (bT2_n) branches as a function of the number of bT1 (bT21_n) branches.
The bT2 branches had a statistically significantly (p < 0.05) larger mean diameter than the bT0 branches (Fig. 6A). The distributions of the mean diameters of the bT0, bT1 and bT2 branches broadly overlapped. The mean diameters of the bT0 and bT1 branches correlated statistically significantly (p < 0.05; Fig. 6B); the same was found for the mean diameters of the bT1 and bT2 branches (p < 0.05; Fig. 6C).
Diameter distributions of branches of the human placenta.
(a–c) Each dot represents the mean diameter of branches of a single isolated villous tree fragment. (a) Mean diameters of branches (md [μm]), separated into various branch terminal distance (bT0, bT1 and bT2). The mean, standard deviation and individual mean diameters (dots) are shown. The mean diameters of branches in position bT2 statistically significantly differ from the mean diameters of branches in position bT0. The p-values and number of villous trees evaluated (n) are given in the graph. (b,c) Correlation of mean diameters of branches in two neighboring hierarchical positions. The regression line (solid line) is shown with a 95% confidence interval (dashed lines). The value of p describing statistically significant deviation of the slope from zero, R2 as a measure of the goodness of fit and the number villous trees evaluated (n) are given in the graphs. (b) The mean diameters of bT1 (bT1_md) branches as a function of the mean diameters of bT0 (bT0_md) branches. (c) The mean diameters of bT2 (bT2_md) branches as a function of the mean diameters of bT1 (bT1_md) branches.
The mean length of the bT0 branches was statistically significantly smaller than the mean length of the bT1 and bT2 branches (p < 0.01; Supplementary Figure S1). The distributions of mean lengths of the bT0, bT1 and bT2 branches broadly overlapped. The length of individual bT0 branches plotted against the angle at which they branch off their respective bT1 branch (planar branching angle of branch bT0) showed a broad distribution of planar branching angles, whereas extremely long branches only occured at planar branching angles below 60° (see Supplementary Figure S1). The 3D datasets of the present study were used as input for a branching analysis, which constisted of a modified Sholl-analysis (for details see methods section and Supplementary Figure S2). This analysis delivered mathematical estimates of branching characteristics, specifically the Sholl-coefficient k and the branching parameter log a (Supplementary Figure S2; Supplementary Table S1).
Means and standard deviations of all 3D parameters are provided in Supplementary Table S1.
Clinical data, tissue processing and histology
Clinical data of the two placenta collections used in the present study were not statistically significantly different from each other (Supplementary Table S2). The histopathological evaluation of HE sections of the placentas analyzed in the present study showed no histopathological deviations. No signs of autolytic change or tissue decomposition were detected. The processing time leading up to availability of raw data for an individual sample was estimated for quantitative analysis on thin 2D histological sections (76.1 h) and Neurolucida-assisted 3D analysis (19.8 h), see Supplementary Table S3. The volume estimates of various tissue components are provided in Supplementary Table S4. The mean volume of the villous tree was determined to be 197.03 ml (+/−48.62 ml standard deviation).
Nonparametric multiple comparison analysis
Nonparametric multiple comparison analysis of the clinical parameters placenta weight (PW), birth weight (BW), fetoplacental weight ratio (PW/BW) and the 3D parameters of the villous tree showed a statistically significant negative correlation (p < 0.05) of PW/BW with the mean planar branching angle of the bT0 branches (Fig. 7A). Accordingly, with increasing PW/BW, the mean planar branching angle of the bT0 branches decreased.
The mean planar branching angle of terminal branches in the human placenta correlates with the fetoplacental weight ratio.
Scatter plots of results of the 3D analysis (a) and analysis of thin 2D histological sections (b,c) against the fetoplacental weight ratio (PW/BW ratio, dimensionless factor). (a) The mean planar branching angles (n = 50) of the bT0 branches (bT0_mpa). The p-value inside the graph was calculated using the multiple nonparametric correlation analysis (Spearman-rho, corrected for multiple comparison according to Benjamini and Hochberg, 199541). Linear correlation analysis (straight line) was calculated with 95% confidence intervals (dashed lines); R2 is a measure for the goodness of linear fit. (b,c) Scatter plots of volume density of the villous tree (y-axis; vdvillous tree; (b)) and volume density of the intervillous space (y-axis; vdintervillous space; (c)) as a function of the PW/BW ratio. Volume densities did not correlate with the PW/BW ratio in the multiple nonparametric correlation analysis.
The volume densities of the villous tree and intervillous space determined by design-based stereology from thin 2D histological sections did not correlate with PW, BW, or PW/BW (Fig. 7B,C). The absolute volumes of the villous tree and intervillous space calculated from the respective volume densities (using placental weight and the average density of placental tissue, for details see methods section) correlated statistically significantly with PW, BW and PW/BW (data not shown). However, it should be noted that these correlations of the absolute volumes with PW, BW and PW/BW are due to usage of PW as a factor in the calculation of the volume density raw data; PW, BW and PW/BW correlate with each other24 and thus also with the absolute volumes.
Discussion
The present study used a novel, quantitative 3D approach to analyze the peripheral parts of villous trees of normal human placentas. In the present study, we show that the 3D analysis of the peripheral human placental villous tree is less laborious than the established analysis strategies (Supplementary Table S3) due to efficient tissue processing and rapid data acquisition. This is enabled by the fact that the Neurolucida-supported 3D analysis is not dependent on any type of pre-existing 3D reconstruction (e.g. from confocal microscopy image stacks or from other sectioning/reconstruction tools), but rather generates its own 3D image during the measurement; the measurement is thus not an extra step. This principle of simultaneous measurement and 3D imaging is thus an elegant and convenient feature of the Neurolucida approach. If the adaptations and precautions of the present study are considered, almost any high quality research microscope equipped for neuronal dendrite tracing will be able to deliver valuable 3D data regarding the villous tree of human placenta. There is no specific requirement for scanning electron microscopy18,25, confocal microscopy, or other sophisticated sectioning/3D reconstruction approaches20,26 in order to obtain such data. This analysis is analogous to the current state of neuroscience, where the majority of the neuronal dendritic and axonal 3D analyses are based on bright-field microscopy7,8,9 due to its broad compatibility with basic histological staining methods.
The 3D analysis in the present study is specifically designed for the analysis of tree structures in 3D. A core feature of this method is the ability to view trees while the trees are simultaneously being reconstructed as a digital replica27. Many details that are invisible on thin 2D histological sections can be analyzed by 3D methods28. Such details include, but are not limited to nodes, branching angles, diameters and lengths of branches in relation to the branching hierarchy/topology. Possible limitations of this method stem from the maximally feasible focus distance of the objective or the villi being too crowded or too thick to be viewed in full depth under a bright-field microscope. With the tracing of villous trees, quantitative microscopic tracing operates at the upper end of the feasible scale. Therefore, the present study focused on the most peripheral part (1–2 mm) of the villous tree of the human placenta. Nevertheless, to the best of our knowledge, no other light microscopic method is able to follow villous trees of the human placenta over an analysis interval of 2 mm with comparable ease and efficiency. Moreover, the focus of this 3D analysis on the most peripheral branches of the villous tree is not necessarily compromising its biological relevance. The peripheral branches of the villous tree arise during the second half of pregnancy2,4,19,29,30,31. Important clinical syndromes, such as preeclampsia and intrauterine growth retardation, also develop during the second half of pregnancy4,32.
The analysis of thin 2D sections/images primarily delivers quantitative data regarding the corresponding 3D space (see, e.g., Supplementary Table S4); thus, the analysis of thin 2D histological sections is highly dependent on the sampling procedures. Using sectioning, the analysis of thin 2D histological sections avoids the potential limitations of optical intransparency or tissue thickness. Aspects other than the volume densities determined in the present study (e.g., star volumes, surface areas, etc.) can also be analyzed on thin 2D histological sections33,34. However, structural details that are invisible in thin 2D histological sections cannot be analyzed in such sections. Notably, nodes, branches, branching angles and branching hierarchy/topology are invisible in thin 2D histological sections (see Fig. 1).
Thus, it is obvious that 3D analysis and the analysis of thin 2D histological sections supplement each other (see also Supplementary Tables S1 and S4). The conventional analysis of thin 2D histological sections can make statistically valid estimates of placental properties. However, such analysis neither produces nor takes into account data describing the 3D branching structure of the villous tree. Three-dimensional analysis performs absolute measurements of nodes, branches and their relations with each other and associated parameters, all of which cannot be done by analysis of thin 2D histological sections.
We found that branches in positions bT0 and bT1 occured at a fixed rate in relation to each other (see Fig. 5B). This finding is self-interpreted because of the general dichotomous branching pattern of the villous tree. However, although the dichotomous branching pattern remains constant, the numbers of branches in positions bT1 to bT2 considerably varied (see Fig. 5C). Taken together, this result would be consistent with the interpretation that branches bT0, bT1 and the nodes connecting these branches form a repetitively occurring 3D building unit of the peripheral villous tree and that these 3D building units are attached to branches in bT2 at a highly variable rate.
Sholl analysis is an important tool for the classification of neurons and their connectivity in the brain35. The strength of this analysis lies in the mathematical recognition of general branching patterns. The present study showed that Sholl analysis can be adapted to the villous tree of the human placenta. Whether Sholl analysis will be able to discriminate between branching patterns of normal placentas and those patterns of complicated pregnancies is beyond the scope of the present study.
For the first time, the present study determined the diameters, lengths and linked parameters, such as surface area and volume, of branches of the peripheral 2 mm of the villous tree, without interference from sectioning artifacts, in many villous trees. Diameters and, to a certain extent, lengths and numbers are principally recognizable as cross-sections of villi in thin 2D histological sections and could potentially be used to reversely identify the terminal/non-terminal position of a sectioned branch. The present study showed statistically significant differences in diameters and lengths from bT0 over bT1 to bT2 (see Figs. 5–7). However, the reverse diagnosis of the terminal or non-terminal position of a given branch by one of these parameters is not feasible due to broad overlap in parameter distributions. Although this is the case, the 3D data of the present study did not suffer from the additional scatter of diameter distributions, which would be caused by the histological sectioning of branches in various angles. Thus, it is not feasible to determine positions of sectioned branches of the most peripheral 2 mm of the villous tree (bT0, bT1 and bT2) on thin 2D histological sections using, e.g., their diameters. This finding is in full agreement with early morphological descriptions of villi in the peripheral part of the villous tree of the human placenta29.
The diameters of branches in each of the positions bT0, bT1 and bT2 showed a linear correlation across these positions in the same villous tree, i.e., thicker branches in position bT2 produced on average thicker branches in positions bT1 and bT1 branches produced on average thicker branches in positions bT0 (see Fig. 6B,C). These diameter relations across branches of the same tree are possibly a tree-specific feature individually characterizing each villous tree.
Branching angles and nodes are structures that are invisible on thin 2D histological sections. These structures “disappear” by the reduction of the 3D tree structure to 2D sections (see Fig. 1) and have never been determined by any known 2D or 3D method of placental analysis. Branching angles and nodes could possibly reflect the different mechanisms of branching that are associated with the villous tree, namely villous sprouting via mesenchymal villi for stem villus branching and angiogenesis-related branching of terminal villi from intermediate villi4. Angiogenesis-related branching of terminal villi from intermediate villi is seen as an especially important mechanism of placental functional adaptation15.
Unexpectedly, in the present study, the branching angles of branches bT0 correlated with the fetoplacental weight ratio PW/BW (Fig. 7A). In contrast, no parameter obtained by the conventional analysis of thin 2D histological sections correlated with the PW/BW ratio (Fig. 7B,C). The PW/BW ratio is a widely used clinical parameter1,4,36. It is outside the scope of the present study to explore possible functional relations between branching angles of terminal branches (bT0) of the villous tree and the PW/BW ratio. However, the plotting of branching angles of terminal branches against the length of terminal branches showed that the branching angles scattered over a wide range and extremely long terminal branches occured at angles below 60° (Supplementary Figure S1). These properties indicated that the terminal branches are much more heterogeneous in structure than previously expected based on the description of terminal villi from histology of thin 2D histological sections2,4,29. A detailed exploration of the structural heterogeneity of terminal branches will likely enable further conceptual understanding of the correlation between the PW/BW ratio and terminal branching angles.
In summary, the present study successfully adapted camera lucida-based analysis strategies to the peripheral part of the villous trees of normal human placentas. Nodes and node-related branching structures (branching hierarchy, angles, diameters and lengths) can now be efficiently measured in 3D. Such primary 3D data can be further processed into secondary parameters by, e.g., Sholl analysis. Branching angles are typical 3D parameters and are correlated with the fetoplacental weight ratio PW/BW, a widely used clinical parameter. The latter encourages the exploration of possible alterations of the 3D structure of villous trees of human placentas from obstetrically complicated pregnancies, particularly preeclampsia and intrauterine growth retardation.
Methods
Study design
The present study used 100 human placentas split into two samples of 50 placentas from clinically normal pregnancies. Both cohorts of placentas were collected at the Department of Obstetrics and Gynecology of the hospital “Dritter Orden”, Munich, Germany. The normal course of each pregnancy was assessed by obstetricians based on clinical information regarding the pregnancy and delivery. Placentas were collected after informed consent of mothers/parents was obtained. Placentas were excluded when: no informed consent of the mothers/parents could be obtained, the language skills of the mothers/parents limited the understanding of information concerning the study, or psychiatric problems or any other condition caused doubts regarding the mothers/parents ability to independently decide. All work was conducted according to relevant guidelines and regulations. This study was approved by the ethics committee of the Ludwig-Maximilians-University (Munich, Germany) under the number 084-11. All data were anonymized. The thickness of the placenta was determined by ultrasound. The placenta weight (PW) was measured prior to tissue sampling and processing without the umbilical cord but with membranes24. The birth weight (BW) was determined immediately after birth and the fetoplacental weight ratio (PW/BW) was calculated1 (Supplementary Table S2).
The cohort of placentas used for the quantitative analysis of thin 2D histological sections was collected between April 2011 and September 2011. The cohort of placentas used for the preparation of single villous trees and subsequent Neurolucida-assisted 3D-analysis was collected between February 2012 and April 2013.
Common procedures for both placenta collections
All placentas were cooled at 4°C immediately after birth and processed within 5.26 h (Median; Max = 23.41 h, Min = 2.31 h; samples for thin 2D histological sections) or 3.28 h (Median; Max = 8.20 h, Min = 1.35 h; samples for 3D analysis) at the Department of Anatomy II of Ludwig-Maximilians-University (Munich, Germany). Six sampling sites were chosen systematically and randomly from each placenta24 and tissue blocks containing the entire thickness of the placenta were collected, fixed and embedded in paraffin as previously described24.
Sections of all specimens were stained with hematoxylin/eosin (HE). All HE sections were qualitatively evaluated in order to confirm the absence of signs of tissue damage/autolysis, inflammation and/or disturbed differentiation of trophoblast.
Two-dimensional thin histological sections
From the six blocks of paraffin-embedded tissue of each placenta, the number 5 was chosen by the roll of a dice. From each placenta, sections of the fifth tissue block were used for the volume estimations on thin 2D histological sections (see Supplementary Table S3).
Whole-mount preparations of villous trees
For the preparation of single, isolated peripheral parts of villous trees (henceforth called “peripheral villous trees”), an additional sample (edge length of 2–3 cm) was collected within a distance of 5 cm from the umbilical cord insertion and transferred to physiological saline at 4°C. The preparation of peripheral villous trees began within one hour after sampling and was performed under a binocular microscope (M400, Wild Heerbrugg, Heerbrugg, Switzerland) with additional light (KL 1500 electronic, Schott/Zeiss, Jena, Germany). Free bushes of peripheral villi were identified, removed using small scissors and fixed in 4.5% formaldehyde (Roti-Histofix, Carl Roth, Karlsruhe, Germany) in small glass jars overnight. The probes were rinsed 3 × 10 minutes with tap water, bleached in 3% H2O2 for 20 minutes and washed 3 × 5 minutes in distilled water. Mayer's hematoxylin was applied for 7 minutes at room temperature with gentle agitation and then the probes were washed in distilled water (acidified by acetic acid to a pH of 2.4) for 3 minutes. Bluing was achieved in tap water for 5 minutes and the probes were then transferred back to distilled water. Following a graded series of ethanol (50%, 70%, 80%, 96% and 100%; 5 min each), the probes were transferred to a mixture of ethanol/xylene (1:1, 5 min) and then to fresh and pure xylene 2 × 5 min. From xylene, the probes were infiltrated with xylene/DPX (1:1, 4 h, DPX: Mountant for histology, Sigma Aldrich nr. 06522, Munich, Germany) and mounted in DPX on a concave slide with enough room to allow the unfolding of the villous tree and such that the 3D structure of the peripheral villous trees remained intact. These preparations have a true 3D character and are free from oppressing contact with the concave slide or cover glass (see Fig. 2).
Analysis of thin 2D histological sections
Volume estimates were performed according to the Cavalieri principle37,38 on single thin (4–6 μm) histological sections using a computerized stereology workstation, which consisted of a modified light microscope (Axioskop; Zeiss, Jena, Germany) with motorized specimen stage for automatic sampling (MBF Bioscience) and stage controller (Type MAC 6000; Ludl Electronics, Hawthorne, NY, USA), focus encoder (Type MT 1271; Heidenhain, Traunreut, Germany), CCD color video camera (1600H × 1200V pixels; MBF Bioscience, Williston, VT, USA) and stereology software (Stereo Investigator version 10; MBF Bioscience). This approach delivers volume densities as raw data, which were allocated to the villous tree (stratified by the villous stroma, vessel lumen, endothelium and syncytiotrophoblast), to the intervillous space and to the fibrinoid. Then, absolute volumes were calculated by multiplying volume densities with the placental volume (placental volume is defined by the placental weight (PW) divided by the density of placental tissue (1.03 g/ml)).
Computer-assisted 3D analysis
Peripheral villous trees were previewed at low magnification (2× microscope objective; Fig. 3A). Tracing with Neurolucida (version 10.54; MBF Bioscience) was performed using a 20× objective (Fig. 3B–Y) with the working direction from the proximal toward the terminal end of the peripheral villous tree (Fig. 3B–Y). Tree ordering for measurements was set to “Terminal Distance Ordering” (Figs. 3B–Y and 2) because the terminal end was the biologically defined end of the isolated peripheral villous trees. Terminal Distance Ordering classifies branches according to their distance in nodes from the terminal end of the villous tree. While viewing the tree, nodes and branches were individually labeled and circumferences of the villi were continuously recorded by mouse wheel functions (Fig. 3B–Y). The measuring system generated a digital 3D replica of the peripheral villous tree under investigation in parallel to the process of tracing. These data were analyzed with Neurolucida Explorer software (MBF Bioscience) using the option “branching structure analysis”. The outcomes can be visualized in various ways in Neurolucida or Neurolucida Explorer software (Fig. 4, Supplementary Movies S1–4).
We used two microscope systems: (i) an Axioskop (Zeiss, Goettingen, Germany) with a motorized XYZ specimen stage (Maerzhaeuser, Wetzlar, Germany), an LEP MAC6000 XYZ 3-axis stage controller (Ludl), a focus encoder (Type MT 1271; Heidenhain) and a color digital camera (3/4″CCD chip 1,92 MP, 1600H × 1200V pixel, MBF Bioscience; and (ii) a BX50 (Olympus, Tokyo, Japan) with motorized XYZ specimen stage (MBF Bioscience), an LEP MAC6000 XYZ 3-axis stage controller (Ludl), focus encoder (Type MT 1271; Heidenhain) and color digital camera (1/2″ CCD chip, 1392 × 1040 pixels, MBF Bioscience).
The parts of villi connecting two nodes or connecting a terminal end with a node were named branches (b). Branches were further classified by their distance to the nearest terminal end (bT, with the T indicating classification by terminal distance). The distance was measured by the number of nodes to the nearest terminal end. Thus, “bT0” encodes a branch in the terminal position, “bT1” encodes a branch one node apart from the nearest terminal position and in general, “bTX” encodes a branch X nodes apart from the nearest terminal end (Fig. 2F). We collected the numbers of nodes and branches on each level of “Terminal Distance Ordering” and data regarding the planar branching angle, diameter, length, surface area and volume of each individual branch. These data were aggregated by the terminal distances of branches for each villous tree as the mean planar branching angle (bTX_mpa), mean diameter (bTX_md), mean length (bTX_ml), mean surface area (bTX_ms) and mean volume (bTX_mv).
The planar branching angle of a given branch was defined as the change in the direction of the branch with respect to the previous branch. The direction of each branch was derived from its endpoints (a node or a terminal end). Of 50 peripheral villous trees, 11 did not show bT2 branches (n = 39 for bT2). Of the remaining 39 bT2 branches, the planar branching angle of bT2 branches could not be determined for six samples due to a missing previous branch (n = 33 for bT2_mpa).
Sholl analysis
Sholl analysis is a mathematical approach for the analysis of dendritic trees39 and Sholl analysis routines are included in software tools designed for tracing dendritic trees, such as Neurolucida27. The tracing direction and analysis direction are usually identical (from perikaryon (root) to terminal end) in the Sholl analysis of dendritic trees. For the placenta, this direction had to be changed because there is only one physiologically defined end of an isolated villous tree; the terminal end. Thus, villous trees were traced from the root (stem villus) to the terminal end, whereas Sholl analysis was manually performed in an opposite direction from terminal end toward the root (see Supplementary Figure S2).
The Sholl analysis of villous trees was performed on the dendrograms exported from Neurolucida Explorer software (see Supplementary Figure S2). The Sholl analysis included the villous trees between the first node (near the terminal end) and the last node (near the root) of the tree under investigation. A grid of measuring lines representing concentric circles was placed in 50 μm intervals from the terminal end to the root. All grid lines falling into the analysis interval between the first and last node were included. The first grid line falling into the analysis interval was set to 1 μm. This setting delivers a set of data linking the distance, in μm and the number of intersecting branches (for details see Supplementary Figure S2). The distances were transformed to the area of the respective concentric circle and the Sholl plot was performed as usual39. The number of intersections with branches by area (N/area) was plotted on the y-axis (logarithmically) whereas distances were plotted on the x-axis (logarithmically). Then, the Sholl regression coefficient (k) was determined by linear regression as the slope of the straight line, together with the y-intercept at X = 0 (log a) in GraphPad Prism software (Version 5.04, GraphPad Software, San Diego, CA, USA), see Supplementary Figure S2.
Statistical analyses
For all investigated parameters (Supplementary Tables S1, S3, S4), the mean and standard deviation were calculated using SPSS software (Version 20, IBM, Armonk, NY, USA)40. Because normal distribution could not be confirmed by Kolmogorov-Smirnov test (SPSS), further statistical analyses were based on non-parametric procedures. Multiple non-parametric bivariate correlation (Spearman-rho) analyses with post-hoc corrections for multiple comparisons according to Benjamini and Hochberg41 (p-value < 0.05) were performed in R42 using the package “psych”43 for multiple comparisons and the function p.adjust for post hoc corrections. Linear regression analyses40 and Kruskal-Wallis tests with Dunn's multiple comparison adjustments were performed in GraphPad software.
References
Boyd, J. D. & Hamilton, W. J. The human placenta. (Heffer, Cambridge, 1970).
Kaufmann, P. Development and differentiation of the human placental villous tree. Bibl. Anat. 22, 29–39 (1982).
Kingdom, J., Huppertz, B., Seaward, G. & Kaufmann, P. Development of the placental villous tree and its consequences for fetal growth. Eur. J. Obstet. Gynecol. Reprod. Biol. 92, 35–43 (2000).
Benirschke, K., Burton, G. & Baergen, R. N. Pathology of the human placenta. (Springer, Berlin and London, 2012).
Barker, S. B., Cumming, G. & Horsfield, K. Quantitative morphometry of the branching structure of trees. J. Theor. Biol. 40, 33–43 (1973).
Wollaston, W. H. Description of the Camera Lucida. Philosophical Magazine 27, 343–347 (1807).
Halavi, M., Hamilton, K. A., Parekh, R. & Ascoli, G. A. Digital reconstructions of neuronal morphology: three decades of research trends. Front. Neurosci. 6, 49–61 (2012).
Marx, M., Günter, R. H., Hucko, W., Radnikow, G. & Feldmeyer, D. Improved biocytin labeling and neuronal 3D reconstruction. Nat. Protoc. 7, 394–407 (2012).
Aguiar, P., Sousa, M. & Szucs, P. Versatile morphometric analysis and visualization of the three-dimensional structure of neurons. Neuroinformatics 11, 393–403 (2013).
Földy, C., Malenka, R. C. & Südhof, T. C. Autism-associated neuroligin-3 mutations commonly disrupt tonic endocannabinoid signaling. Neuron 78, 498–509 (2013).
Eisenstein, M. Neural circuits: Putting neurons on the map. Nature 461, 1149–1152 (2009).
Helmstaedter, M., Sakmann, B. & Feldmeyer, D. The relation between dendritic geometry, electrical excitability and axonal projections of L2/3 interneurons in rat barrel cortex. Cereb. Cortex 19, 938–950 (2009).
Oberlaender, M. et al. Three-dimensional axon morphologies of individual layer 5 neurons indicate cell type-specific intracortical pathways for whisker motion and touch. Proc. Natl. Acad. Sci. USA 108, 4188–4193 (2011).
Oberlaender, M. et al. Cell type-specific three-dimensional structure of thalamocortical circuits in a column of rat vibrissal cortex. Cereb. Cortex 22, 2375–2391 (2012).
Kaufmann, P., Mayhew, T. M. & Charnock-Jones, D. S. Aspects of human fetoplacental vasculogenesis and angiogenesis. II. Changes during normal pregnancy. Placenta 25, 114–126 (2004).
Howard, C. V. & Reed, M. G. Unbiased stereology: Three-dimensional measurement in microscopy. (BIOS Scientific Publ., Oxford, 1998).
Mayhew, T. M. Stereology and the placenta: where's the point? – a review. Placenta 27, S17–25 (2006).
Castellucci, M., Scheper, M., Scheffen, I., Celona, A. & Kaufmann, P. The development of the human placental villous tree. Anat. Embryol. (Berl) 181, 117–128 (1990).
Habashi, S., Burton, G. J. & Steven, D. H. Morphological study of the fetal vasculature of the human term placenta: scanning electron microscopy of corrosion casts. Placenta 4, 41–56 (1983).
Karen, P., Jirkovska, M., Tomori, Z., Demjenova, E., Janacek, J. & Kubinova, L. Three-dimensional computer reconstruction of large tissue volumes based on composing series of high-resolution confocal images by GlueMRC and LinkMRC software. Microsc. Res. Tech. 62, 415–422 (2003).
Jirkovska, M. et al. Topological properties and spatial organization of villous capillaries in normal and diabetic placentas. J. Vasc. Res. 39, 268–278 (2002).
Jirkovska, M., Janacek, J., Kalab, J. & Kubinova, L. Three-dimensional arrangement of the capillary bed and its relationship to microrheology in the terminal villi of normal term placenta. Placenta 29, 892–897 (2008).
Jirkovska, M. et al. The branching pattern of villous capillaries and structural changes of placental terminal villi in type 1 diabetes mellitus. Placenta 33, 343–351 (2012).
Haeussner, E., Schmitz, C., Koch, F. V. & Frank, H. G. Birth weight correlates with size but not shape of the normal human placenta. Placenta 34, 574–582 (2013).
Burton, G. J. The fine structure of the human placental villus as revealed by scanning electron microscopy. Scanning Microsc. 1, 1811–1828 (1987).
Kubínová, L. et al. Confocal stereology and image analysis: methods for estimating geometrical characteristics of cells and tissues from three-dimensional confocal images. Physiol. Res. 53 Suppl 1, S47–55 (2004).
Glaser, J. & Glaser, E. Neuron imaging with neurolucida — A PC-based system for image combining microscopy. Comput. Med. Imag. Grap. 14, 307–317 (1990).
van Oppenraaij, R. H. F. et al. Vasculogenesis and angiogenesis in the first trimester human placenta: An innovative 3D study using an immersive virtual reality system. Placenta. 30, 220–222 (2009).
Kaufmann, P., Sen, D. K. & Schweikhart, G. Classification of human placental villi. I. Histology. Cell Tissue Res. 200, 409–423 (1979).
Sen, D. K., Kaufmann, P. & Schweikhart, G. Classification of human placental villi. II. Morphometry. Cell Tissue Res. 200, 425–434 (1979).
Mayhew, T. M. Fetoplacental angiogenesis during gestation is biphasic, longitudinal and occurs by proliferation and remodelling of vascular endothelial cells. Placenta 23, 742–750 (2002).
Rath, W. Geburtshilfe und Perinatologie. (Thieme, Stuttgart, 2010).
Karimu, A. L. & Burton, G. J. Star volume estimates of the intervillous clefts in the human placenta: How changes in umbilical arterial pressure might influence the maternal placental circulation. J. Dev. Physiol. 19, 137–142 (1993).
Mayhew, T. M. & Sisley, I. Quantitative studies on the villi, trophoblast and intervillous pores of placentae from women with well-controlled diabetes mellitus. Placenta 19, 371–377 (1998).
Radley, J. J. et al. Chronic behavioral stress induces apical dendritic reorganization in pyramidal neurons of the medial prefrontal cortex. Neuroscience 125, 1–6 (2004).
Cetin, I. & Taricco, E. [Clinical causes and aspects of placental insufficiency]. The Placenta and Human Developmental Programming [Burton G., Barker D., Moffett A., & Thornburg K. (eds.)] [114–125] (Cambridge University Press, Cambridge, 2011).
Gundersen, H. J. & Jensen, E. B. The efficiency of systematic sampling in stereology and its prediction. J. Microsc. 147, 229–263 (1987).
Schmitz, C. & Hof, P. R. Design-based stereology in neuroscience. Neuroscience 130, 813–831 (2005).
Sholl, D. A. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 87, 387–406 (1953).
Bronstein, I. N. & Semendjajew, K. A. Taschenbuch der Mathematik (Deutsch, Thun, 2001).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 57, 289–300 (1995).
R Core Team. R: A Language and Environment for Statistical Computing. Vienna and Austria: R Foundation for Statistical Computing. URL: http://www.R-project.org/ (Date of access:12/04/2014) (2013).
Revelle, W. psych: Procedures for Psychological, Psychometric and Personality Research. Evanston and Illinois: Northwestern University. URL: http://CRAN.R-project.org/package=psych (Date of access:12/04/2014) (2013).
Acknowledgements
The authors acknowledge the skillful technical assistance and diligent work of the entire team of technicians of the Department of Anatomy II at LMU Munich, namely B. Aschauer, A. Baltruschat, U. Fazekas, S. Kerling, C. Stanglmair, B. Mosler and S. Tost. We would also like to express our thanks to the obstetricians, midwives and nurses of the Dritter Orden hospital (Munich, Germany) who enabled the clinical work of this study with great care and engagement. Benedikt Kropp assisted with R graphics code. P. Angstman assisted in video compression. For proofreading and helpful discussions regarding the preparation of the manuscript, we would like to thank N. Angstman, N. Csaszar, S. Milz and K. Sternecker. Three-dimensional analysis is part of the doctoral thesis of EH. The analysis of thin 2D histological sections is part of the doctoral thesis of AB.
Author information
Authors and Affiliations
Contributions
E.H. performed the 3D analysis. A.B. performed analysis on thin 2D histological sections. F.v.K., C.S. and H.G.F. designed the study. F.v.K. was responsible for obstetrical tasks. E.H. and H.G.F. prepared figures and tables. H.G.F., C.S., F.v.K. and E.H. wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Materials
Supplementary Information
Supplementary Movie S1
Supplementary Information
Supplementary Movie S2
Supplementary Information
Supplementary Movie S3
Supplementary Information
Supplementary Movie S4
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Haeussner, E., Buehlmeyer, A., Schmitz, C. et al. Novel 3D Microscopic Analysis of Human Placental Villous Trees Reveals Unexpected Significance of Branching Angles. Sci Rep 4, 6192 (2014). https://doi.org/10.1038/srep06192
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06192
This article is cited by
-
Placental Morphology Is Associated with Maternal Depressive Symptoms during Pregnancy and Toddler Psychiatric Problems
Scientific Reports (2018)
-
Whole organ vascular casting and microCT examination of the human placental vascular tree reveals novel alterations associated with pregnancy disease
Scientific Reports (2017)
-
Novel 3D light microscopic analysis of IUGR placentas points to a morphological correlate of compensated ischemic placental disease in humans
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.