Abstract
The molecular program controlling hematopoietic differentiation is not fully understood. Here, we describe a family of zebrafish genes that includes a novel hematopoietic regulator, draculin-like 3 (drl.3). We found that drl.3 is expressed in mesoderm-derived hematopoietic cells and is retained during erythroid maturation. Moreover, drl.3 expression correlated with erythroid development in gata1a- and spi1b-depleted embryos. Loss-of-function analysis indicated that drl.3 plays an essential role in primitive erythropoiesis and, to a lesser extent, myelopoiesis that is independent of effects on vasculature, emergence of primitive and definitive progenitor cells and cell viability. While drl.3 depletion reduced gata1a expression and inhibited erythroid development, enforced expression of gata1a was not sufficient to rescue erythropoiesis, indicating that the regulation of hematopoiesis by drl.3 extends beyond control of gata1a expression. Knockdown of drl.3 increased the proportion of less differentiated, primitive hematopoietic cells without affecting proliferation, establishing drl.3 as an important regulator of primitive hematopoietic cell differentiation.
Similar content being viewed by others
Introduction
Vertebrate hematopoiesis occurs in successive waves that originate from distinct anatomical regions in the developing embryo1,2. In zebrafish, primitive granulocytes arise predominantly from the anterior lateral mesoderm (ALM) whereas primitive erythrocytes arise exclusively from the posterior lateral mesoderm (PLM), which later becomes the intermediate cell mass (ICM)3,4,5,6. Subsequently, a transient wave of definitive cells with myeloid and erythroid potential (EMPs), form in the tail posterior blood island (PBI) between 24–40 hours post-fertilization (hpf)7. Overlapping with this wave, by 28 hpf the hemogenic endothelium in the aorta-gonad-mesonephros (AGM) region gives rise to definitive hematopoietic stem cells (HSC)8,9,10,11,12,13. Cell tracing experiments demonstrated that primitive granulocytes are present through 3 days post-fertilization (dpf) and cell structure analysis showed that primitive erythrocytes can be detected at 4 dpf, indicating the persistence of primitive cells despite the emergence of definitive waves4,6,14.
While the spatiotemporal production of hematopoietic cells is well described, the molecular circuitry controlling this process continues to unfold15,16. In vertebrate embryos, scl and lmo2 are expressed at the earliest stages of hematopoietic specification and are essential for the generation of primitive hematopoietic lineages17,18,19,20. Thus, scl or lmo2 depleted zebrafish embryos display decreased expression of erythroid gata1a and myeloid spi1b in mesoderm-derived cells19,20,21,22,23 and certain markers of more mature cells22,23. In contrast, runx1 and cmyb are key regulators of definitive HSC development10,24. There are a number of transcriptional regulators that direct hematopoietic lineage specification/differentiation, cell proliferation and/or survival25,26,27,28. Within this framework, gata1a is essential for primitive erythropoiesis while spi1b is necessary for primitive myelopoiesis, although cross-antagonism between these regulators also contribute to cell fate outcomes29,30,31,32. To better understand the regulation of hematopoietic differentiation, we used zebrafish to examine still elusive components of this regulatory network.
Here, we report the discovery of three new genes, drl.1, drl.2 and drl.3, that exhibit very high homology to the draculin (drl) gene and are predicted to encode zinc-finger proteins of, so far, unknown function. Drl was previously identified in a whole mount RNA in situ hybridization (WISH) screen of zebrafish cDNA libraries (http://zfin.org/)33. We examine the role of the drl gene family in zebrafish embryonic and hematopoietic development and focus on one family member, drl.3, that is specifically required for primitive hematopoietic differentiation.
Results
Identification of the draculin-like gene family
Basic Local Alignment Search (BLAST) of the drl gene to the zebrafish genome identified three draculin-like genes. Drl, drl.1, drl.2 and drl.3 are clustered consecutively on chromosome 5. All four genes contain three exons with very high homology in coding and non-coding exon sequences, with drl.3 having the longest coding sequence of the group (Supplementary Fig. S1a). Drl, Drl.1 and Drl.2 contain 13 consecutive Cys2-His2 (C2H2) zinc-finger domains, while Drl.3 contains 20 C2H2 domains (Supplementary Fig. S1b). Multiple adjacent C2H2 motifs are known to confer DNA binding activity, which suggests a role for these factors as transcriptional modulators34,35. Consistent with this idea, a Drl.3-specific antibody detected the protein in nuclear and cytoplasmic lysates from zebrafish embryos (Supplementary Fig. S1c).
To examine the conservation of the drl family between species, we performed BLASTp analysis of Drl.3 protein to non-redundant protein sequences in various metazoans. The phylogenetic relationship between homologous proteins shows segregation into species-specific clusters, but not protein-specific clusters (Supplementary Fig. S2a). The zebrafish drl genomic region is similar to a region on chicken chromosome 19, but is not syntenic to the human or mouse genomes (Supplementary Fig. S2b). The genes flanking the drl cluster are located on human 17 and murine 11 chromosomes, however 6 of the 21 genes examined have not been identified in chicken (Supplementary Fig. S2c). While clear drl family homologs were not identified, the human genes encoding proteins homologous to Drl.3 are clustered to several distinct chromosomal regions (Supplementary Fig. S2d), supporting the idea that the zebrafish drl cluster has a corresponding cluster in the human genome. As the functions of the human genes are largely unknown and many of the corresponding mouse homologs have not been determined (Supplementary Fig. S2e), understanding drl family activity will likely guide the identification of their functional equivalents in higher vertebrates.
Expression analysis of drl family members
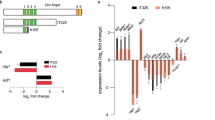
To determine the expression profile of drl family members, we performed reverse transcription-PCR (RT-PCR) from a series of developmentally staged embryos (Fig. 1a). The primers were gene-specific, with two exceptions: first, primers for drl.2 also amplified drl DNA (Supplementary Fig. S1d) and, second, drl primers amplified an additional upper band from total RNA, which was sequenced and determined to be drl.1 (Fig. 1a). Our results from embryo samples revealed that drl family genes are not maternally expressed. Expression was detected by 30% epiboly through 120 hours post-fertilization (hpf). Semi-quantitative RT-PCR analysis of drl family members in 14 somite stage embryos showed varied levels of expression, with drl.2 and drl.3 having the highest and lowest levels of expression, respectively (Fig. 1b). While high homology between drl, drl.1 and drl.2 prohibits the real-time quantification of these genes, this method could be used to analyze drl.3 expression. Real-time PCR determined that drl.3 expression was highest at 30% epiboly and progressively decreased through 48 hpf (Fig. 1c). This may reflect a progressive restriction in cell type-specific expression during development or a decrease in cellular expression levels during embryogenesis.
Drl gene family members are expressed throughout embryonic development and in multiple hematopoietic lineages.
(a) RT-PCR analysis of the drl family members and β−actin from pooled embryos at the indicated ages. (b) Semi-quantitative (q) RT-PCR of drl genes and β-actin from pooled 14 somite stage embryos (left panel). Arrows indicate the bands that were used for quantitation, which is shown in the right panel. Compared to drl, *P = 0.0150, **P = 0.0052, , xP = 0.0360,
, xP = 0.0360, , *P = 0.0142, **P = 0.0029 and ***P < 0.001 (Student's t-test). (c) Quantitative real-time PCR analysis of drl.3 in whole embryos at the indicated ages. The relative expression of drl.3 was normalized to the expression of gapdh. (d–e) WISH of drl.2 (d) and drl.3 (e) during embryonic development. Ages of embryos are indicated. Embryos at 2-cell and 30% epiboly stages are shown in a lateral view, animal pole at the top. 50% epiboly embryos are shown in an animal pole view, dorsal to the right. Embryos at 75% and 90% epiboly are shown as lateral views, dorsal to the right. Staged embryos at 10 somites, 18 somites, 24 and 48 hpf are shown as lateral views, anterior to the left. White arrows indicate ALM; asterisks indicate ICM; black arrows indicate PLM; arrowheads indicate AGM cells. Insets show magnified AGM region of corresponding embryo. (f) RT-PCR analysis of the drl family members and β−actin from purified populations of hematopoietic cells. The age of the embryos and the transgenic lines from which cells were purified are indicated. (g) Quantitative real-time PCR analysis of drl.3 in 24 hpf sorted hematopoietic cells. The relative expression of drl.3 was normalized to the expression of gapdh. ***P < 0.0001 (Student's t-test). (b–c, g) Bars show mean ± S.D. (a–b, f) Full-length gel images are provided in Supplemental Figure 8.
, *P = 0.0142, **P = 0.0029 and ***P < 0.001 (Student's t-test). (c) Quantitative real-time PCR analysis of drl.3 in whole embryos at the indicated ages. The relative expression of drl.3 was normalized to the expression of gapdh. (d–e) WISH of drl.2 (d) and drl.3 (e) during embryonic development. Ages of embryos are indicated. Embryos at 2-cell and 30% epiboly stages are shown in a lateral view, animal pole at the top. 50% epiboly embryos are shown in an animal pole view, dorsal to the right. Embryos at 75% and 90% epiboly are shown as lateral views, dorsal to the right. Staged embryos at 10 somites, 18 somites, 24 and 48 hpf are shown as lateral views, anterior to the left. White arrows indicate ALM; asterisks indicate ICM; black arrows indicate PLM; arrowheads indicate AGM cells. Insets show magnified AGM region of corresponding embryo. (f) RT-PCR analysis of the drl family members and β−actin from purified populations of hematopoietic cells. The age of the embryos and the transgenic lines from which cells were purified are indicated. (g) Quantitative real-time PCR analysis of drl.3 in 24 hpf sorted hematopoietic cells. The relative expression of drl.3 was normalized to the expression of gapdh. ***P < 0.0001 (Student's t-test). (b–c, g) Bars show mean ± S.D. (a–b, f) Full-length gel images are provided in Supplemental Figure 8.
To address this question, we used whole mount RNA in situ hybridization (WISH). Drl expression is widespread at 30% epiboly, is expressed in developing mesoderm tissue during gastrulation (50%–90% epiboly) and becomes restricted to hematopoietic cells during somitogenesis (data not shown)33. WISH indicated that the expression patterns of drl.1 (data not shown), drl.2 and drl.3 (Fig. 1d and e, respectively) are identical to drl. Markers of early hematopoietic cells are expressed in the ALM and PLM/ICM of 10 and 18 somite stage embryos2; at these stages drl family members are expressed exclusively in these hematopoietic regions. At 24 hfp, drl family transcripts were detected in developing erythroid cells and very weakly in myeloid cells. Expression at 48 hpf was detected in erythroid cells and cells in the aorta-gonad-mesonephros (AGM) region.
Using drl.3 as an example, we examined whether this gene is expressed in the hemogenic endothelium in the ventral aorta (VDA), the AGM region that gives rise to definitive runx1-expressing HSC. Transverse sections determined that drl.3 was expressed in cells within the VDA in 28 hpf embryos, similar to runx1-expressing cells (Supplementary Fig. S3a). Double WISH analysis of 36 hpf embryos showed that a portion of the runx1+ cells in the VDA coexpress drl.3 (Supplementary Fig. S3b–c), indicating that drl.3 is expressed in definitive stem or/and progenitor cells.
Next, we performed RT-PCR analysis of purified hematopoietic cells (Fig. 1f). Flow cytometric purification of the hematopoietic populations was based on the transgenic expression of fluorescent labels controlled by the promoters marking the vascular/hemangioblasts (fli1), myeloid progenitor cells (spi1), erythroid progenitor cells (gata1), mature myeloid cells (mpx), definitive HSC and thrombocytes (itga2b) and lymphoid cells (rag2 and lck). Drl gene family transcripts were detected in all of these cell types, with several exceptions. To determine if a lack of a band was due to RT-PCR failure, genes that were not detected in a sample were retested for expression in those cell types. Initial evaluation did not detect drl.3 in 2 dpf itga2b:GFP+ and 5 dpf fli1:GFP+ (Fig. 1f). However, two additional independent experiments clearly detected drl.3 in these samples (Supplementary Fig. S1e and data not shown), indicating that drl.3 is expressed in these cells. In contrast, triplicate independent analyses failed to detect drl.1 expression in 5 dpf itga2b:GFP+ cells (Fig. 1f, Supplementary Fig. S1e and data not shown), suggesting that drl.1 is not expressed in this cell type at this age. Quantitative real-time PCR analysis showed that drl.3 has significantly higher expression in gata1:RFP+ cells than cells sorted from fli1:GFP, spi1:GFP and mpx:GFP 24 hpf embryos (Fig. 1g), indicating that drl.3 expression is retained during primitive erythropoiesis. Further studies are needed to quantify the cell-type-specific levels of drl, drl.1 and drl.2.
Since drl.3 is expressed in runx1+ cells during embryogenesis, we examined whether drl family members are expressed in adult hematopoietic cells. RT-PCR analysis of gata1:RFP+ transgenic cells from adult zebrafish detected expression of each drl family member (Supplementary Fig. S3d), although the drl.2 band may reflect drl.2 and/or drl expression. Next, we purified mature myeloid mpx:GFP+ from adult kidney marrow to compare to gata1:RFP+ cells (Supplementary Fig. S3g–h). Murine Scl and Gata1 expression increase as cells differentiate into erythroid progenitors and decrease during myeloid specification36,37,38, whereas expression of mammalian Spi1 increases during myeloid differentiation37,39. The gata1:RFP+ cells were separated into large and small size groups based on the prediction that the larger cells would be less differentiated with lower levels of scl and gata1a expression. Indeed, scl and gata1a expression was detected in both gata1:RFP+ cell size pools, but was significantly lower in the larger gata1:RFP+ cells compared to the smaller cells (Supplementary Fig. S3g, P = 0.0003 and P = 0.0097, respectively). This expression pattern suggests that the small cell fraction is more differentiated than the larger cell fraction, although both likely contain a mix of cells. Similar levels of spi1b and drl.3 expression were detected in both gata1:RFP+ populations (Supplementary Fig. S3g). In contrast, drl.3 and gata1a had higher levels of expression in erythroid lineage cells than in mature myeloid mpx:GFP+ cells (Supplementary Fig. S3h; LC P = 0.0115, SC P = 0.0221), similar to the expression pattern of drl.3 during embryonic hematopoiesis.
The draculin gene family regulates embryonic patterning
To gain insights into the roles of drl family members in embryogenesis, we performed gene knockdown experiments using antisense morpholinos. The target sites are diagrammed in Fig. 2a. The translation blocking MO2 was used in a previous study, but its effects were not carefully tested40. The morpholino sequences predict that (1) MO2 should inhibit the protein translation of all 4 family members while (2) MO3 should inhibit splicing of drl, drl.1 and drl.2 without affecting drl.3; thus, MO3 is likely to have overlapping or similar effects as MO2. In contrast, (3) L3MO should disrupt splicing of drl.3 specifically without altering the processing of other family member transcripts. Consistent with these expectations, RT-PCR analysis of RNA from 24 hpf morpholino and mismatch control-injected individual embryos (morphants) determined that splice site-directed MO3 decreased drl, drl.1 and drl.2 transcripts by 67% (P = 0.0382), 100% (P < 0.0001) and 97% (P = 0.0088), respectively, but did not affect drl.3 transcripts (P = 0.4588) (Fig. 2b). Conversely, the splice-site morpholino specifically targeting drl.3 (L3MO) significantly decreased this transcript (80%, P = 0.0064) without affecting other family members (Fig. 2b). Detection of endogenous Drl.3 protein showed that MO2 and L3MO decreased Drl.3 protein in 24 and 48 hpf morphant lysates (Fig. 2c–d).
Knockdown of the drl gene family causes developmental defects.
(a) (Left panel) Diagram comparing exon homology and showing the location of morpholino-targeted sequences. The percent homology for each exon compared to drl is indicated. (Right panel) Alignment of morpholino target sequence (in red) compared to corresponding sequence in each drl family member. DNA base pairs are color-coded. (b) (Upper panel) Individual embryo RT-PCR analysis of drl family members in control and morpholino-injected embryos as indicated. MM indicates a 5 base pair mismatch control morpholino. UI = uninjected. (Lower panels) Quantitation of drl gene family transcripts from RT-PCR analysis. (Left) Control (MO2MM) versus MO3 samples. (Right) Control (L3MM) compared to L3MO samples. Samples were normalized to β-actin. Rel. = Relative. Bars show mean ± S.D. from triplicates; *P = 0.0382, **P = 0.0088, ***P < 0.0001 and  (Student's t-test). (c) Detection of Drl.3 protein and Tubulin in 24 and 48 hpf lysates extracted from uninjected (UI), L3MO- or MO2-injected embryos, 293T cells and 293T cells expressing Drl.3. (d) Quantitation of Drl.3 from Western blot analysis (from c, normalized to Tubulin). Norm. rel. = normalized relative; AU = arbitrary units. (e) Bright field microscopy of uninjected, control morpholino-injected embryos (MO2MM and L3MM) and morpholino injected-embryos at 18 somites. Lateral views, head to the left, dorsal upward. (f) WISH of scl (dark blue), krox20 (red, rhombomeres 3 and 5) and myoD (red, somites) in 9–10 somite stage embryos. Dorsal views of flat mounted embryos, anterior to left. (e–f) The number of the embryos with the representative phenotype per total number of embryos is indicated for each panel. Full-length gel images and western exposures for the cropped panels (b–c) are shown in Supplemental Figure 8.
(Student's t-test). (c) Detection of Drl.3 protein and Tubulin in 24 and 48 hpf lysates extracted from uninjected (UI), L3MO- or MO2-injected embryos, 293T cells and 293T cells expressing Drl.3. (d) Quantitation of Drl.3 from Western blot analysis (from c, normalized to Tubulin). Norm. rel. = normalized relative; AU = arbitrary units. (e) Bright field microscopy of uninjected, control morpholino-injected embryos (MO2MM and L3MM) and morpholino injected-embryos at 18 somites. Lateral views, head to the left, dorsal upward. (f) WISH of scl (dark blue), krox20 (red, rhombomeres 3 and 5) and myoD (red, somites) in 9–10 somite stage embryos. Dorsal views of flat mounted embryos, anterior to left. (e–f) The number of the embryos with the representative phenotype per total number of embryos is indicated for each panel. Full-length gel images and western exposures for the cropped panels (b–c) are shown in Supplemental Figure 8.
Simultaneous knockdown of multiple drl family genes with either MO2 or MO3 resulted in the same developmental defects, including decreased ventrally-derived tail tissues and a shortened body axis at 18 hpf (Fig. 2e). Knockdown of bloody fingers (blf) has been shown to cause similar morphological defects40. The Blf zinc finger protein is similar to Drl and Drl.3 (Supplementary Fig. 4a), but RT-PCR analysis showed that the drl morpholinos do not alter blf expression levels (Supplementary Fig. 4b–c). WISH analysis of MO2- and MO3-injected embryos at 9–10 somites revealed convergence/extension defects as indicated by the increased lateral extension of krox20 marking rhombomeres 3 and 5, myoD labeled somites and decreased anterior-posterior expanse of scl expression (Fig. 2f). MO2 and MO3 morphants appear dorsalized at 72 and 48 hpf, with some variation in the severity of the phenotype (Supplementary Fig. S5a–b and data not shown). A higher morpholino dose correlated with a higher proportion of embryos displaying severe or moderate loss of tissue and body axis truncation at 24 hpf, whereas lower doses resulted in fewer affected embryos (Supplementary Fig. S5c). This suggests that the morpholino effects are specific and establishes a role for this family in the development of ventral tissue during zebrafish embryogenesis. Depletion of drl.3 alone did not cause morphological defects (Fig. 2e–f).
Drl.3 is required for erythropoiesis
Since drl.3 is strongly expressed in the (gata1a+) erythroid lineage, we examined whether drl.3 is essential for erythropoiesis. We found that 40–47% of 24 hpf drl.3 morphants (3 WISH experiments, 218 total embryos) had decreased numbers of gata1a+ cells (Fig. 3a). Manual counting of gata1a+ or o-dianisidine stained hemoglobin+ cells determined that morphants with decreased erythropoiesis had on average about half the number of cells as outwardly normal morphants (Supplementary Fig. S5d–e). In the studies below, affected embryos have obviously decreased erythroid cell numbers that are estimated to be ≤60% of normal. Consistent with gata1a deficiency, mature erythroid marker hbae1 was decreased in 23–51% of 24 hpf drl.3 morphants (186 total embryos; triplicate experiments) compared to controls (Fig. 3a). In contrast, at 24 hpf, expression of the fli1:EGFP transgene was normal in drl.3 morphants (Fig. 3a), indicating that drl.3 is not required for vasculogenesis.
Drl.3 is essential for erythroid development.
(a) Control- (mismatch, L3MM) and drl.3 morpholino- (L3MO) injected embryos at 24 hpf showing live Tg(fli1a:EGFP) marking of the vasculature and gata1a and hbae1 WISH, as indicated. (b) Embryos at 35 hpf showing live Tg(fli1a:EGFP) pattern, with a magnified view of the PBI region and o-dianisidine stained embryos with an enlargement of the anterior region. (c) RT-PCR analysis of individual Tg(gata1:DsRED) control and L3MO-injected embryos. Drl.3 morphants were sorted for normal or decreased (estimated ≤ 60% of normal) numbers of circulating erythrocytes at 48 hpf. Full-length gel images for these cropped panels are provided in Supplemental Figure 8. (d) Quantitation of the levels of drl.3 expression normalized to β-actin from RT-PCR analysis in (c). **P = 0.0019 and **P = 0.0015 (Student's t-test). (e) WISH of gata1a and slc4a1a in 48 hpf control and drl.3 morphants. (f) O-dianisidine stained control and drl.3 morphants at 4 dpf; lateral views (left panels) and ventral views of the anterior region (right panels). (a–b, e–f) The number of the embryos with the representative phenotype per total number of embryos is shown; lateral views with head to the left, dorsal upward.
At 35 hpf, drl.3 morphants with decreased erythrocytes in circulation had normal fli1:EGFP expression despite showing slight edema in the posterior blood island (PBI) (Fig. 3b). The PBI edema (8–15% in triplicates; 167 total embryos) was only present in embryos with a severe decrease in numbers of circulating cells, as assessed by bright field microscopy (data not shown). Similarly, gata1:RFP transgenic drl.3 morphants showed that 8–10% of the embryos had decreased erythroid cells in circulation (data not shown). Consistent with an ablation of mature erythroid cells, 16–25% of drl.3 morphants (triplicate; 213 total embryos) showed decreased numbers of o-dianisidine+ cells (Fig. 3b). RT-PCR analysis revealed that drl.3 morphants with normal or decreased gata1:RFP+ circulating cells had comparable levels of drl.3 knockdown (Fig. 3c–d), suggesting that drl.3 knockdown gives rise to an erythroid phenotype that is not fully penetrant. By 48 hpf, expression of gata1a was indistinguishable between drl.3 morphants and controls at 48 hpf (Fig. 3e) while drl.3 morphants continued to display decreased erythroid slc4a1a+ (Fig. 3e; 51/63 embryos) and o-dianisidine+ cells (Supplementary Fig. S5e; 22/65 embryos). The numbers of o-dianisidine+ cells remained low in 4 dpf drl.3 morphants (Fig. 3f; 16/132 embryos), suggesting that primitive erythropoiesis does not recover.
Monteiro et al. established that the genetic interactions of gata1a are different in primitive versus definitive hematopoietic cells41, a concept that is likely to apply to other factors such as Drl.3. It remains unclear why slc4a1a WISH at 48 hpf showed a higher percentage of embryos that were affected by drl.3 knockdown compared to o-dianisidine staining. Nonetheless, our findings show that drl.3 is important for the generation of erythrocytes during primitive erythropoiesis. Drl family morphants also display decreased o-dianisidine stained cells at 2 dpf (Supplementary Fig. S5f–g). The percent of embryos with severe and moderate decreases in erythrocytes was dependent on the morpholino dose and the severity of the erythroid phenotype correlated with the severity of the morphological defects (Supplementary Fig. S5h–i). This suggests that decreased erythropoiesis in MO2 and MO3 morphants are likely secondary to the developmental abnormalities, although the possibility remains that the drl genes contribute to erythropoiesis.
Drl.3 deficiency impairs primitive myelopoiesis
Since drl.3 deficiency inhibits erythroid lineage development, we postulated that drl.3 functions at a very early stage of hematopoiesis. Scl precedes gata1a expression in the PLM and scl is essential for the derivation of all embryonic hematopoietic lineages19,42. Figure 1f shows that scl expression is normal in 9–10 somite stage drl.3 morphants. Drl.3 morphants continued to show a normal pattern of scl expression at 24 and 48 hpf (Fig. 4a). At 48 hpf, definitive hematopoietic stem/progenitor cell marker runx1 was easily detected in AGM cells even when erythroid slc4a1a expression was decreased (Fig. 4a). These results indicate that drl.3 is not essential for the emergence of primitive hematopoietic progenitors or definitive stem cells.
Knockdown of drl.3 transiently decreases myeloid cells without altering the emergence of primitive progenitor and definitive stem cells.
(a) WISH of scl at 24 and 48 hpf and runx1 (dark blue)/slc4a1a (red) at 48 hpf in L3MM- and L3MO-injected embryos, as labeled. Embryos shown as lateral views. (b) WISH of spi1b, l-plastin and mpx at 24 hpf and l-plastin and mpx 48 hpf. Dorsal, anterior (24 hpf only, right panels) and lateral views are shown. (c) Quantitation of the number of the WISH spi1b+, l-plastin+ and mpx+ cells in the anterior of the embryo at 24 hpf and total body l-plastin+ and mpx+ cells at 48 hpf in L3MM (blue) and L3MO-injected (red) embryos (N = 8 for each column except for mpx at 24 hpf where N = 15, bars show mean ± S.E.). **P = 0.0044, ***P < 0.0001 and *P = 0.0156 (Student's t-test). (d) Quantitative real-time PCR analysis of drl.3 in whole embryo RNA samples from 24 and 48 hpf drl.3 morphants (red) and controls (L3MM, blue, set to 1, arbitrary units). **P = 0.0042 and ***P < 0.0001 (Student's t-test). (e) Quantitative real time PCR analysis of scl, spi1b, gata1a, gata1b and drl.3 in whole embryo RNA samples from pools of 30 hpf drl.3 morphants (red) compared to control-injected embryos (blue, set to 1, arbitrary units). *P = 0.0114, **P = 0.0083 and ***P = 0.0005 (Student's t-test). (d–e) Bars show mean ± S.D., from three independent experiments. Expression was normalized to gapdh. (f) Tail region of spi1:GFP embryos at 54 hpf, 6 hours after tail transection. Selected L3MO embryos had decreased circulating cells; controls were randomly selected and had normal circulation. Red boxes = tail cut region. (g) Number of spi1:GFP+ cells in tail cut region in control or drl.3 morphants. P = 0.007 (Student's t-test). (h) WISH of lysozymeC/lysC (blue) and slc4a1a (red) at 54 hpf, 6 hours after tails were cut. (i) Number of lysC+ cells in an equal sized region surrounding the tail in the indicated embryos at 54 hpf, 6 hours after tail cuts were performed. P = 6.77E-5, Student's t-test.
As drl.3 expression was detected in myeloid cells, we questioned whether drl.3 depletion would impact myelopoiesis. At 24 hpf, drl.3 deficiency resulted in decreased numbers of spi1b-expressing primitive myeloid lineage cells in 40–43% of the morphants (triplicate; 175 total embryos) (Fig. 4b–c) and l-plastin-expressing monocytes/macrophages in 45–49% of the morphants (triplicate; 168 total embryos, data not shown). Codetection of lysozymeC (lysC) and hbae1 expression revealed that 24 hpf anemic morphants (22%) had significantly fewer lysC+ granulocytes compared to controls (Fig. 4b–c). Similarly, 24 hpf drl.3 morphants displayed decreased numbers of myeloperoxidase (mpx)-expressing granulocytes in 76% of the morphants (Fig. 4b–c). However, 48 hpf drl.3 morphants had normal numbers of l-plastin-, mpx- and lysC-expressing myeloid cells (Fig. 4b–c). Real-time PCR analysis determined that drl.3 transcripts were decreased in morphants by 78% (P < 0.0001) and 56% (P = 0.0042) at 24 and 48 hpf, respectively (Fig. 4d).
To confirm the WISH results, we performed quantitative real-time PCR of RNA from pools of 30 hpf drl.3 morphants and control-injected embryos. Drl.3 expression was severely decreased in morphants, which was expected since L3MO disrupts the derivation of the transcript detected in this assay (Fig. 4e). Spi1b and gata1b expression levels were significantly decreased in drl.3 morphants (Fig. 4e). The expression levels of scl and gata1a were not affected by drl.3 knockdown at this time point (Fig. 4e). WISH of gata1a is normal in 48 hpf drl.3 morphants, consistent with the real time quantitation of gene expression.
Myeloid cells accumulate over time at sites of acute injury in zebrafish embryos43,44. To evaluate the myeloid inflammatory response, tail transections were performed in 48 hpf spi1:GFP and non-transgenic embryos and live GFP+ or WISH lysC+ cells were quantified in equal sized areas surrounding the injury 6 hours later. Spi1:GFP drl.3 morphants were visually selected based on low numbers of cells in circulation; control-injected embryos were randomly selected and had normal circulation. Drl.3 morphants with decreased circulation or slc4a1a expression had significantly fewer injury-localized GFP+ (Fig. 4f–g, P = 0.007) and lysC-expressing (Fig. 4h–I, P = 6.8E-5) cells, respectively, compared to control embryos. In contrast, gata1a morphants, which lack erythrocytes and have increased lysC+ cells at 2 dpf, had a slight, but not significant increase in lysC+ cells near the acute injury site. These data suggest that an absence of primitive erythrocytes does not necessarily block the myeloid cell response to injury and that drl.3 morphant myeloid cell function is compromised despite the recovery of normal numbers of l-plastin-, mpx- and lysC-expressing cells.
Forced expression of drl does not compensate for drl.3 deficiency
WISH analysis of 24 hpf drl.3 morphants showed decreased numbers of cells expressing the four drl genes in the ICM and anterior hematopoietic regions compared to the control-injected embryos (Supplementary Fig. S6a), ruling out increased drl family member expression as a compensatory mechanism in drl.3 morphants. To determine that inhibition of hematopoiesis is specifically due to knockdown of drl.3, we rescued the L3MO-induced defects by injecting drl.3 mRNA (Fig. 5a–c). Enforced expression of drl.3 significantly reduced the proportion of drl.3 morphants with decreased gata1a+ (L3MO = 36%; L3MO + drl.3 mRNA = 9%; P < 0.0001), slc4a1a+ (L3MO = 49%; L3MO + drl.3 mRNA = 13%; P < 0.0001) and l-plastin+ (L3MO = 46%; L3MO + drl.3 mRNA = 27%; P = 0.0445) cells, confirming that the defects in erythropoiesis and myelopoiesis are caused by drl.3 deficiency. Although there is extremely high homology between drl family members and slightly decreased expression of other family members in drl.3 morphants (Fig. 2b and Supplementary Fig. S6a), enforced expression of drl did not compensate for the loss of drl.3 (Supplementary Fig. S6b). Thus, drl.3 and drl have non-redundant, or at least partially non-overlapping, functions.
L3MO-induced defects are due to knockdown of drl.3 activity.
(a) WISH of gata1a at 24 hpf, l-plastin at 25 hpf and slc4a1a at 48 hpf in L3MM- and L3MO-injected embryos and embryos co-injected with L3MO and drl.3 mRNA. Embryos shown as lateral views, head left. WISH for l-plastin also shows dorsal, anterior views. (b) Percent of L3MM- and L3MO-injected and L3MO/drl.3 mRNA co-injected embryos with normal (wt) or decreased (*; estimated ≤ 60% of normal) numbers of gata1a or slc4a1a-expressing cells. The numbers of embryos are indicated in the columns. ***P < 0.0001 (Fisher's exact test). (c) Percent of embryos with normal (wt) or decreased (*) numbers of l-plastin-expressing cells, as labeled. The numbers of embryos are indicated in the columns. *P = 0.0445 (Fisher's exact test).
Knockdown of gata1a and spi1b affect drl.3 expression
Since spi1 and gata1 play key roles in the development of myeloid and erythroid lineages31,32,45, respectively, we were curious about the genetic relationship of drl.3 with these other factors. To address this question, we morpholino-silenced gata1a and spi1b and assessed drl gene family expression (Fig. 6a). At 24 hpf, gata1a morphants showed a loss of slc4a1a+ cells, increased numbers of anterior l-plastin+ cells and a robust spi1b+ population in the ICM while spi1b morphants showed decreased numbers of l-plastin-expressing cells (Fig. 6a). These data are consistent with published studies showing that Gata1a and Spi1b control primitive hematopoietic cell fate and confirm the activity of these morpholinos25,29,30,31,46. Turning to the drl family, 24 hpf gata1a morphants displayed decreased drl family-expressing cells in the ICM and anterior hematopoietic region compared to control embryos (Fig. 6a–b). As drl family expression closely tracks with the derivation of erythroid cells, this result is likely due to the altered ICM cell composition in gata1a morphants. However, it remains to be determined whether Gata1a regulates transcription of the drl family genes, which could also underscore these findings. The numbers of cells expressing drl family members was increased in the anterior hematopoietic region of spi1b morphants (Fig. 6a–b), consistent with the acquisition of erythroid features by spi1b-deficient anterior hematopoietic cells32,41.
Loss of gata1a and spi1b affect drl gene family expression.
(a) WISH of drl family members in gata1a and spi1b morphants (MO) compared to uninjected embryos at 24 hpf. From top to bottom: drl, drl.1, drl.2, drl.3, l-plastin (dark blue)/slc4a1a (red) and spi1b. Lateral views, head to the left (left panels); anterior, dorsal views (right panels). The number of the embryos with the representative phenotype out of the total number of embryos is indicated. Arrows in the panels showing dorsal views indicate an increase or decrease in the numbers of WISH+ cells. (b) Quantitation of the number of drl gene family-expressing cells in the anterior hematopoietic region of control (UI), gata1a morphants and spi1b morphants at 24 hpf. N = 10 for each column. Bar shows mean ± S.D. *P = 0.0013, **P = 0.0002,  and ***P ≤ 0.0001 (Student's t-test). (c) WISH of drl family members in the indicated embryos at 48 hpf. Embryos shown as lateral views. Horizontal arrows indicate the region where cells in circulation can be visualized. Downward facing arrows indicate decreased WISH+ cell numbers. (d) Percent of L3MM-injected, L3MO-injected and L3MO/gata1a mRNA co-injected embryos that have normal (blue, wt) or low numbers of erythroid cells (red, *estimated ≤ 60% of normal) based on o-dianisidine staining at 48 hpf. (e) Quantitative analysis of uninjected, gata1a MO-injected and gata1a MO/drl.3 mRNA co-injected embryos that have normal (blue, wt) or low numbers of erythroid cells (red, *) based on o-dianisidine staining at 48 hpf. (d–e) Numbers of embryos are indicated in the columns. Statistical significance was analyzed using Fisher's exact test.
and ***P ≤ 0.0001 (Student's t-test). (c) WISH of drl family members in the indicated embryos at 48 hpf. Embryos shown as lateral views. Horizontal arrows indicate the region where cells in circulation can be visualized. Downward facing arrows indicate decreased WISH+ cell numbers. (d) Percent of L3MM-injected, L3MO-injected and L3MO/gata1a mRNA co-injected embryos that have normal (blue, wt) or low numbers of erythroid cells (red, *estimated ≤ 60% of normal) based on o-dianisidine staining at 48 hpf. (e) Quantitative analysis of uninjected, gata1a MO-injected and gata1a MO/drl.3 mRNA co-injected embryos that have normal (blue, wt) or low numbers of erythroid cells (red, *) based on o-dianisidine staining at 48 hpf. (d–e) Numbers of embryos are indicated in the columns. Statistical significance was analyzed using Fisher's exact test.
At 48 hpf, gata1a morphants show an absence of slc4a1a-expressing cells and increased numbers of l-plastin-expressing cells (Fig. 6c and published studies32,47). Gata1a morphants displayed decreased drl family-expressing cells in hematopoietic pools on the yolk, which are likely erythrocytes, compared to controls (Fig. 6c). Spi1b morphants had decreased numbers of l-plastin-expressing cells, as expected32; however, at this time point, drl family expression appeared normal (Fig. 6c). These data suggest that gata1a promotes the derivation or survival of drl family-expressing cells or transcription of these genes, while spi1b knockdown transiently impacts the expression pattern of drl family members during primitive hematopoiesis. Additional studies are necessary to establish the transcriptional versus non-transcriptional control Gata1a exerts on drl family expression.
Enforced expression of gata1a mRNA did not alter the proportion of drl.3 morphants with defective erythropoiesis (Fig. 6d, P = 0.6282). Similarly, we determined that 98% of both gata1a morphants and gata1a morphants co-injected with drl.3 mRNA had decreased o-dianisidine+ erythrocytes (Fig. 6e, P = 1.000). These results demonstrate that enforced expression of drl.3 or gata1a is not sufficient to rescue the defects caused by depletion of the other gene, suggesting that either they do not have a linear relationship to one another and/or represent only a portion of the differentiation program that they control.
Drl.3 is important for hematopoietic cell differentiation
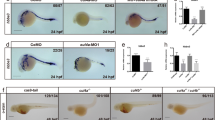
Acridine orange staining and immunodetection of cleaved Caspase-3 revealed that drl.3 deficiency does not alter cell survival at 24 hpf (Supplementary Fig. S7a–b), suggesting that other cellular mechanisms underlie the loss of maturing hematopoietic populations. Using phosphorylated histone H3 (pH3) as a mitotic indicator, we found no difference in the percent of mitotic cells in the ICM of control and drl.3 morphants (Supplementary Fig. S7c). Gata1a and pu.1b mark cells destined to become erythroid and myeloid, respectively, partly due to the expression of these genes as well as the distinct regional localization of the cells. The gata1:RFP and spi1:GFP transgenes mark primitive hematopoietic progenitors even when endogenous gene expression is depleted32. Fluorescence-activated cell sorting (FACS) analysis showed no difference in the percent of gata1:RFP+ cells from 24 hpf drl.3 morphants (2.89%) and control embryos (2.17%) (Fig. 7a). Furthermore, we did not detect significant differences in the percent of spi1:GFP+ cells at 24 hpf (control 6.91%, L3MO 8.94%; P = 0.3083) (Supplementary Fig. S8a–b) or fli1a:GFP+ cells at 30 hpf (control 14.70%, L3MO 16.50%; P = 0.8473) (Supplementary Fig. S8c). Together, these findings indicate that survival and proliferation are normal in drl.3-deficient hematopoietic cells.
Drl.3 is necessary for hematopoietic cell differentiation.
(a) FACS plots of 24 hpf cells from wild-type uninjected (UI) and Tg(gata1:DsRED) embryos injected with L3MM or L3MO as indicated. (b) Quantitation of the percent of large cells RFP+ populations from FACS analysis of L3MM- (blue) and L3MO-injected (red) Tg(gata1:DsRED) embryos, as indicated. *P = 0.0107; **P = 0.0021 (Chi-Squared test). (c) FACS plots of 48 hpf cells from wild-type uninjected (UI) and Tg(gata1:DsRED) embryos injected with L3MM and L3MO as indicated. (a, c) The percent of RFP+ cells out of total cells and the percent of large (FSC high) and smaller sized cell (FSC low) populations are indicated. (d) May-Grunwald-Giemsa (MGG) staining of purified Tg(gata1:DsRED) cells from the indicated 24 hpf morphants. *p = less differentiated progenitor cell; p = progenitor cell, more differentiated. (e) Cell type distribution in purified Tg(gata1:DsRED) cells at 24 hpf based on MGG staining. L3MM versus L3MO (*p and p cells), P = 0.0004 (Fisher's exact test). (f–g) MGG staining of purified 48 hpf Tg(gata1:DsRED) cells (f) and quantitation of MGG cell type distribution (g). p = progenitor cells; e = erythroid cells (≤8 μm); m = myeloid; l = lymphoid. L3MM versus L3MO (e and p cells), P < 0.0001 (Fisher's exact test). The yellow-blue color balance of merged L3MM and L3MO images was slightly adjusted in Photoshop. (e, g) The cell count for each cell type is indicated in the appropriate column. (h) FACS plots of cells from 24 hpf Tg(spi1:EGFP) L3MM- (top) and L3MO-(bottom) injected embryos. An arrow indicates the less mature population (P = 0.0069, Chi-squared test). Supplementary Figure S6a–b shows gating for GFP+ cells. (i) Real-time PCR of scl, spi1b, gata1a, gata1b and drl.3 in purified large and small size populations of gata1:RFP+ cells from 24 hpf L3MM- and L3MO-injected embryos. The relative expression in small L3MM cells was set to 1, arbitrary units. Expression was normalized to gapdh. Bars show mean ± S.D., triplicate experiments. **P = 0.0083, ***P = 0.0004 and ***P = 0.0009 (Student's t-test).
Erythroid cell size decreases during maturation. Using forward light scatter as an indicator of cell size, we found an increased proportion of gata1:RFP+ large cells (16.98%) in 24 hpf drl.3 morphants compared to the control population (9.72%) (Fig. 7a–b). At 48 hpf, the drl.3 morphant cell population continued to be skewed towards larger cells (Fig. 7b–c). May-Grundwald-Giemsa (MGG) staining of purified 24 hpf gata1:RFP cells distinguished cells with large nuclei, diffuse chromatin and a cell size that is larger, which is consistent with a less differentiated state compared to the smaller, more granular cells (Fig. 7d). From 24 hpf embryos, we found 34% (148 cells counted) of the drl.3-deficient cells appeared less differentiated compared to 14% (105 cells counted) of cells from control-injected embryos (P < 0.0001) (Fig. 7d–e). Similarly, drl.3-deficient 48 hpf gata1:RFP cells showed an increased proportion of less differentiated cells (Fig. 7f–g). The altered composition of the hematopoietic population in drl.3 morphants was also detected in cells purified from spi1:GFP and fli1a:EGFP transgenic embryos at 24 and 30 hpf, respectively (Fig. 7h, Supplementary Fig. S8c, d–e). As myeloid cells become large and very granular during differentiation, we used the forward (FSC, size) and side scatter (SSC, granularity) distribution of spi1:GFP+ cells to distinguish between more or less differentiated cells (Fig. 7h and Supplementary Fig. S8f–g). Recovery of the myeloid lineage was observed by these methods, as the scatter distribution was normal for 48 hpf drl.3 morphant spi1:GFP+ cells (Supplementary Fig. S8f–g).
To better define the gata1:RFP large and small cell fractions, we quantified expression of a small set of hematopoietic genes. Murine scl and gata1 expression initially increase during erythroid differentiation, whereas spi1b expression decreases37,38. The expression patterns of these genes indicate that the large cells are less differentiated than the smaller cells (Fig. 7i). Drl.3 was easily detected in both control populations, with the control large cells displaying high drl.3 expression (Fig. 7i). The drl.3 morphant large cells had slightly decreased gata1a and gata1b expression compared to control samples, however the larger cells expressed comparatively less gata1a than the smaller cells (Fig. 7i). This suggests that the decrease in gata1a+ cells detected by WISH is primarily due to an altered hematopoietic composition. Interestingly, the differential level of scl and gata1a expression was more robust in adult samples compared to the embryonic populations and drl.3 and spi1b had similar levels in adult (large and small) erythroid lineage cells (Supplementary Fig. S3i and Fig. 7i). It is unclear whether the different expression levels/patterns in primitive versus adult samples are due to differences in the cell-type composition of the purified populations or/and different levels of gene expression in these samples. Nonetheless, these data suggest that drl.3 deficiency causes a block in the differentiation of primitive erythroid cells.
Discussion
We have identified new zebrafish genes, drl.1, drl.2 and drl.3, that are highly homologous to drl. Knockdown of drl, drl.1 and drl.2 together result in convergence/extension defects, however silencing of drl.3 uncovered a specific requirement for this gene in hematopoiesis. The relatively low expression levels of drl.3 in embryos compared to other family members raise the possibility that dose may be important for their roles. Enforced expression of drl is not sufficient to rescue drl.3 morphants, supporting the idea that drl.3 activity is not entirely redundant with other family members and highlighting the functional divergence of drl.3. These studies do not rule out the possibility that drl, drl.1 and drl.2 may contribute to hematopoiesis. Likewise, drl.3 may contribute to embryonic patterning, although ablation of drl.3 alone is not sufficient to induce defects in non-hematopoietic tissues. Moreover, expression in definitive stem/progenitors and adult cells raises the possibility that drl.3 and perhaps other family members, participate in the production of hematopoietic cells throughout the lifespan of zebrafish.
The drl gene family encodes proteins with multiple consecutive C2H2 zinc-finger domains, an architecture that has been shown to direct sequence-specific DNA binding activity34,35. Given the unique requirement for drl.3 in hematopoiesis compared to other family members, we postulate that the 7 additional zinc-finger domains that are only present in Drl.3 and/or the unique amino- and carboxyl-terminal regions may underlie its hematopoietic-specific function. Alternately, the different levels of drl family expression may correlate to their different developmental roles. Further studies are necessary to better understand the molecular activity of Drl.3. Moreover, given the conservation of the hematopoietic regulatory program and the presence of C2H2 gene clusters in the human genome, it is likely that a human equivalent of drl.3 exists, although additional studies are needed to identify this gene/s.
Drl.3 deficiency severely compromised erythropoiesis while myelopoiesis was only modestly affected. Drl.3 is highly expressed in erythroid lineage cells compared to myeloid lineage cells, which may explain the sensitivity of erythropoiesis to drl.3 knockdown. The mechanism underlying the partial penetrance of the erythroid defect remains unclear. Possible explanations for the transient decrease in myelopoiesis are that (1) primitive myelopoiesis is delayed but recovers, perhaps by activating intrinsic compensatory mechanisms and/or that mesoderm-derived cells are not absolutely dependent on drl.3 for myeloid differentiation, (2) primitive myelopoiesis is rescued by definitive hematopoiesis and/or (3) cell non-autonomous signals promote myeloid cell homeostasis. It remains to be determined whether, given time, the myeloid inflammatory response to injury would recover in drl.3 morphants.
Embryonic knockdown of zebrafish gata1a and spi1b alters erythroid and myeloid lineage specification32,41,45, respectively, whereas depletion of drl.3 disrupts both the myeloid and erythroid lineages. Enforced expression of gata1a and drl.3 could not rescue the erythroid defects due to knockdown of the other gene. These data suggest that drl.3 and gata1a govern distinct aspects of erythropoiesis.
Drl.3 knockdown increases the proportion of less differentiated primitive erythroid-lineage cells and possibly primitive myeloid-lineage cells and causes a corresponding decrease of mature myeloid and erythroid cells during primitive hematopoiesis without affecting mesoderm specification, the emergence of mesoderm-derived progenitors or HSC, cell proliferation and viability. The patterns of scl and runx1 expression suggest that the normal controls over of the regulation of these genes are engaged despite the block in cell differentiation. Nevertheless, our data suggest that drl.3 is important for the homeostatic balance between undifferentiated cells and mature, functional cells during hematopoietic development.
Methods
Zebrafish maintenance
Wild-type AB stocks of Danio rerio and the transgenic lines Tg(gata1:dsRED), Tg(fli1:EGFP), Tg(spi1:EGFP), Tg(mpx:EGFP), Tg(rag2:EGFP) and Tg(lck:EGFP) were raised and maintained under standard laboratory conditions. The FCCC IACUC approved all zebrafish procedures.
Cloning of drl family genes
The zebrafish drl genes were PCR-amplified from a zebrafish cDNA library with gene-specific primers (Supplementary Table S1). Full-length coding sequences were cloned into TOPO-TA (Invitrogen) then subcloned into the EcoRI site of the pCS2+ vector and confirmed by DNA sequencing. The accession numbers of drl, drl.1 and drl.3 are NM_130977, JX844126, JX844127, respectively (Supplementary Table S1). Drl.2 cDNA sequence is in Supplementary Fig. S1. The Ensembl designators are listed in Supplementary Table S1.
Microinjections and whole mount RNA in situ hybridization (WISH)
Gene Tools, LLC (http://www.gene-tools.com/) designed the morpholinos. Sequences are provided in Table S1. Published gata1a and spi1b morpholinos were used as previously described32. Morpholinos (1–4 ng per embryo) were injected into the yolk of 1-cell stage embryos, except L3MO, which was heated at 50°C for 3–5 minutes and then injected into the cell of 1-cell stage embryos. To prepare mRNAs, drl, drl.3 and gata1a clones in pCS2+ were linearized by digestion with NotI (high fidelity, NEB) and in vitro transcribed with the mMessage mMachine SP6 Kit (Ambion). mRNAs were purified using NucAway Spin Columns (Ambion) and injected into the cell of 1-cell stage embryos.
All 4 drl antisense riboprobes were prepared by using BstBI (NEB) and T3 RNA polymerase (Ambion), while all sense riboprobes were prepared by using ApaI (NEB) and SP6 RNA polymerase (Ambion). RNA probes were generated with digoxigenin or fluorescein RNA labeling mix (Roche) and purified by NucAway Spin Columns according to the manufacturer protocols. Digoxigenin- or fluorescein-labeled antisense riboprobes for zebrafish scl, krox20, myoD, gata1a, hbae1, slc4a1a, spi1b, l-plastin and runx1 were synthesized according to published literature3,32,43,48,49,50,51,52. Embryo fixation and WISH were performed as previously described32. Vector Labs BCIP/NBT substrate kit and Roche Fast Red Tablets were used to detect alkaline phosphatase conjugated secondary antibodies (Roche). WISH embryos were embedded in Optimum cutting temperature (Tissue-Tek OCT, Sakura Finetek) freezing medium for 8-micron thick tranverse cryosections.
O-dianisidine staining and cell quantitation
Hemoglobin was detected by incubating live embryos in o-dianisidine (Sigma) + H2O2 as previously described53. To determine the threshold for whether whole mount embryos are scored as having normal or decreased numbers of erythroid cells, we used control and drl.3 morphants analyzed by gata1a WISH at 24 hpf and o-dianisidine staining at 48 hpf. We then scored the embryos as having apparently normal or low numbers of erythroid cells by microscopy. From images of each embryo, we counted the numbers of gata1a+ cells within a matching 3-somite region of the trunk and the o-dianisidine+ cells on comparable regions of the yolk surface. The average number of cells in embryos observed to have decreased erythropoiesis was about half that of apparently normal embryos, although there was some variation (Supplementary Fig. S4e–f). Based on these data, we can estimate that embryos visually scored as having decreased erythropoiesis have ≤60% the normal numbers of cells.
FACS analysis and cytospin
For each embryonic sample, 100–200 dechorionated transgenic embryos were pooled, washed with ice-cold FACS buffer (10 mM HEPES, 5% FBS, deficient RPMI), mechanically dissociated and filtered through an 85 μm mesh. Whole kidneys from 15 wild-type AB strain adult fish (31 months old), 6 gata1:dsRED adults (15–21 months old) and 6 mpx:EGFP adults (13 months old) were mechanically dissociated in ice-cold FACS buffer and filtered through an 85 μm mesh. Propidium iodide (PI) was added to preparations from GFP transgenics. Up to 100,000 fluorescently labeled cells were collected from each sample using Becton-Dickinson FACS Vantage SE or Aria II cell sorters. Sorted cells were stained with May-Grunwald-Giemsa stain as previously described53.
RT-PCR and quantitative real-time PCR
Reverse Transcription-PCR (RT-PCR) analysis of drl family gene expression was performed on RNA extracted with TRIzol (Invitrogen) from individual or pools of 10–20 embryos. RNA was extracted from sorted hematopoietic populations using a phenol:chloroform:isoamyl alcohol protocol54 and DNAse treatment (Ambion). The primers for these studies are listed in Table S1. For quantitative real-time PCR, cDNA was prepared from whole embryo or purified cell RNA extracts with Super-Script II First Strand Synthesis System (Invitrogen) and oligo-dT primers (Ambion). Gel images were acquired from a UVP DigiDoc-It Imaging system with a Canon Powershot G9 camera and Canon Utilities Remotecapture DC 3 software. Quantitative real-time PCR was performed using the TaqMan Master Mix (Applied Biosystems) according to the manufacturer protocols. Oligonucleotide sequences were designed and provided by Applied Biosystems. Gene expression changes were normalized to gapdh.
Drl.3 antibody production and immunodetection
A rabbit polyclonal antibody was generated in the FCCC animal facility against Drl.3 carboxyl-terminal peptide sequence CEGEHDQTSLLKG (Alpha Diagnostic International). Antibody was purified from 6 combined bleeds of rabbit serum using the Sulfolink Immobilization Kit for Peptides (Pierce) according to manufacturer's instructions, except 0.5% Acetic Acid with 150 mM sodium chloride (NaCl) was used as elution buffer. Anti-Drl.3 was diluted 1∶25 in 5% milk in PBST (Fisher Scientific) and incubated overnight at 4°C for western blots. Monoclonal Tubulin antibody (Sigma) was used at a 1∶5000 dilution. Peroxidase conjugated secondary antibodies (Jackson Immunoresearch Labs Inc) and ECL Substrate (Pierce) were used for detection.
HEK-293T cells were cultured using standard procedures (www.ATCC.org) and transfected with pcDNA3.1+ or a construct encoding a FLAG-Drl.3 fusion protein using Lipofectamine 2000 (Invitrogen). Cells were lysed with 1% Triton X-100, 150 mM NaCL, 20 mM HEPES, 5 mM EDTA, 5 mM sodium floride, 0.2 mM sodium orthovanadate and cOmplete protease inhibitor cocktail (1 tablet/10 mL buffer, Roche). Whole embryo lysates were prepared by pooling 20 randomly selected embryos per sample, removing the yolk using mechanical dissociation and lysing in 20 mM HEPES, 150 mM NaCl, 1% deoxycholate, 1% Nonidet P-40, 1% SDS with cOmplete protease inhibitor cocktail (Roche). Cytoplasmic lysates were prepared from dissociated embryos lysed in 10 mM HEPES, 10 mM potassium chloride, 1.5 mM magnesium chloride (MgCl2), 1 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail and 10% Nonidet P-40. Following centrifugation, the cytoplasmic fraction (supernatant) was collected. The nuclear fraction (pellet) was lysed using 20 mM HEPES, 400 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 5% glycerol and protease inhibitor cocktail (Roche). Standard procedures were used for 10% polyacrylamide electrophoresis and transfer of proteins to nitrocellulose55.
Whole mount immunofluorescent detection was performed as previously described56. Anti-cleaved-Caspase 3 (BD Biosciences) and anti-phospho-Histone H3 (Santa Cruz) were diluted 1∶100 and 1∶200, respectively. Alexa-488-conjugated secondary antibodies (Life Technologies) were diluted 1∶200 dilution. Samples were mounted for fluorescence microscopy in Vectashield mounting medium with DAPI (Vector Labs).
Imaging
Bright field images were captured on a Nikon SMZ 1500 microscope with a Spot Insight camera using Spot Basic software. Fluorescent and cytospin images were captured on a Nikon SMZ 1500 microscope with a Nikon Digital Sight camera or a Nikon Eclipse 80i microscope with a CRi Nuance Multispectral camera and a Nikon Digital Sight camera. Images were obtained using NIS Element AR 3.0 software or CRi Nuance 2.10.0 software. Composite images of different focal planes were compiled in Adobe Photoshop without altering the characteristics of the images. Confocal images were acquired on a Nikon Eclipse TE-2000E/C1 laser scanning confocal microscope using EZ-C1 3.80 software (Nikon) and analyzed with Fiji software (distributed by ImageJ, General Public License).
References
Carroll, K. J. & North, T. E. Oceans of Opportunity: Exploring Vertebrate Hematopoiesis in Zebrafish. Exp Hematol, 10.1016/j.exphem.2014.05.002 (2014).
Davidson, A. J. & Zon, L. I. The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene. 23, 7233–7246 (2004).
Bennett, C. M. et al. Myelopoiesis in the zebrafish, Danio rerio. Blood. 98, 643–651 (2001).
Le Guyader, D. et al. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood. 111, 132–141 (2008).
Ransom, D. G. et al. Characterization of zebrafish mutants with defects in embryonic hematopoiesis. Development. 123, 311–319 (1996).
Weinstein, B. M. et al. Hematopoietic mutations in the zebrafish. Development. 123, 303–309 (1996).
Bertrand, J. Y. et al. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 134, 4147–4156, 10.1242/dev.012385 (2007).
Bertrand, J. Y. et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 464, 108–111, 10.1038/nature08738 (2010).
Burns, C. E. et al. Isolation and characterization of runxa and runxb, zebrafish members of the runt family of transcriptional regulators. Exp Hematol. 30, 1381–1389 (2002).
Kalev-Zylinska, M. L. et al. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 129, 2015–2030 (2002).
Kissa, K. & Herbomel, P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 464, 112–115, 10.1038/nature08761 (2010).
Ma, D., Zhang, J., Lin, H. F., Italiano, J. & Handin, R. I. The identification and characterization of zebrafish hematopoietic stem cells. Blood. 118, 289–297, 10.1182/blood-2010-12-327403 (2011).
Thompson, M. A. et al. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 197, 248–269 (1998).
Belair, C. D., Peterson, R. E. & Heideman, W. Disruption of erythropoiesis by dioxin in the zebrafish. Dev Dyn. 222, 581–594, 10.1002/dvdy.1213 (2001).
Gieger, C. et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 480, 201–208, 10.1038/nature10659 (2011).
Novershtern, N. et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 144, 296–309, 10.1016/j.cell.2011.01.004 (2011).
Porcher, C. et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 86, 47–57 (1996).
Yamada, Y. et al. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci U S A. 95, 3890–3895 (1998).
Dooley, K. A., Davidson, A. J. & Zon, L. I. Zebrafish scl functions independently in hematopoietic and endothelial development. Dev Biol. 277, 522–536 (2005).
Patterson, L. J. et al. The transcription factors Scl and Lmo2 act together during development of the hemangioblast in zebrafish. Blood. 109, 2389–2398 (2007).
Gering, M., Yamada, Y., Rabbitts, T. H. & Patient, R. K. Lmo2 and Scl/Tal1 convert non-axial mesoderm into haemangioblasts which differentiate into endothelial cells in the absence of Gata1. Development. 130, 6187–6199, 10.1242/dev.00875 (2003).
Patterson, L. J., Gering, M. & Patient, R. Scl is required for dorsal aorta as well as blood formation in zebrafish embryos. Blood. 105, 3502–3511, 10.1182/blood-2004-09-3547 (2005).
Juarez, M. A., Su, F., Chun, S., Kiel, M. J. & Lyons, S. E. Distinct roles for SCL in erythroid specification and maturation in zebrafish. J Biol Chem. 280, 41636–41644, 10.1074/jbc.M507998200 (2005).
Soza-Ried, C., Hess, I., Netuschil, N., Schorpp, M. & Boehm, T. Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proc Natl Acad Sci U S A. 107, 17304–17308, 10.1073/pnas.1004640107 (2010).
Gangenahalli, G. U. et al. Stem cell fate specification: role of master regulatory switch transcription factor PU.1 in differential hematopoiesis. Stem Cells Dev. 14, 140–152, 10.1089/scd.2005.14.140 (2005).
Lukin, K., Fields, S., Hartley, J. & Hagman, J. Early B cell factor: Regulator of B lineage specification and commitment. Semin Immunol. 20, 221–227, 10.1016/j.smim.2008.07.004 (2008).
Kaushansky, K. Historical review: megakaryopoiesis and thrombopoiesis. Blood. 111, 981–986, 10.1182/blood-2007-05-088500 (2008).
Ruscetti, F. W., Akel, S. & Bartelmez, S. H. Autocrine transforming growth factor-beta regulation of hematopoiesis: many outcomes that depend on the context. Oncogene. 24, 5751–5763, 10.1038/sj.onc.1208921 (2005).
Liew, C. W. et al. Molecular analysis of the interaction between the hematopoietic master transcription factors GATA-1 and PU.1. J Biol Chem. 281, 28296–28306, 10.1074/jbc.M602830200 (2006).
Rekhtman, N. et al. PU.1 and pRB interact and cooperate to repress GATA-1 and block erythroid differentiation. Mol Cell Biol. 23, 7460–7474 (2003).
Rekhtman, N., Radparvar, F., Evans, T. & Skoultchi, A. I. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 13, 1398–1411 (1999).
Rhodes, J. et al. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 8, 97–108 (2005).
Herbomel, P., Thisse, B. & Thisse, C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 126, 3735–3745 (1999).
Pabo, C. O., Peisach, E. & Grant, R. A. Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem. 70, 313–340, 10.1146/annurev.biochem.70.1.313 (2001).
Wolfe, S. A., Nekludova, L. & Pabo, C. O. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 29, 183–212, 10.1146/annurev.biophys.29.1.183 (2000).
Suzuki, N. et al. Identification and characterization of 2 types of erythroid progenitors that express GATA-1 at distinct levels. Blood. 102, 3575–3583, 10.1182/blood-2003-04-1154 (2003).
Akashi, K., Traver, D., Miyamoto, T. & Weissman, I. L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404, 193–197, 10.1038/35004599 (2000).
Iwasaki, H. et al. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity. 19, 451–462 (2003).
Chen, H. M. et al. Neutrophils and monocytes express high levels of PU.1 (Spi-1) but not Spi-B. Blood. 85, 2918–2928 (1995).
Sumanas, S., Zhang, B., Dai, R. & Lin, S. 15-zinc finger protein Bloody Fingers is required for zebrafish morphogenetic movements during neurulation. Dev Biol. 283, 85–96 (2005).
Monteiro, R., Pouget, C. & Patient, R. The gata1/pu.1 lineage fate paradigm varies between blood populations and is modulated by tif1gamma. EMBO J. 30, 1093–1103, 10.1038/emboj.2011.34 (2011).
Chen, A. T. & Zon, L. I. Zebrafish blood stem cells. J Cell Biochem (2009).
Zhang, Y. et al. In vivo interstitial migration of primitive macrophages mediated by JNK-matrix metalloproteinase 13 signaling in response to acute injury. J Immunol. 181, 2155–2164 (2008).
Mathias, J. R. et al. Live imaging of chronic inflammation caused by mutation of zebrafish Hai1. J Cell Sci. 120, 3372–3383, 10.1242/jcs.009159 (2007).
Nerlov, C., Querfurth, E., Kulessa, H. & Graf, T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 95, 2543–2551 (2000).
Su, F. et al. Differential regulation of primitive myelopoiesis in the zebrafish by Spi-1/Pu.1 and C/ebp1. Zebrafish. 4, 187–199, 10.1089/zeb.2007.0505 (2007).
Galloway, J. L., Wingert, R. A., Thisse, C., Thisse, B. & Zon, L. I. Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev Cell. 8, 109–116 (2005).
Song, H. D. et al. Hematopoietic gene expression profile in zebrafish kidney marrow. Proc Natl Acad Sci U S A. 101, 16240–16245, 10.1073/pnas.0407241101 (2004).
Liu, T. X. et al. Dominant-interfering C/EBPalpha stimulates primitive erythropoiesis in zebrafish. Exp Hematol. 35, 230–239 (2007).
Stewart, R. A. et al. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev Biol. 292, 174–188, 10.1016/j.ydbio.2005.12.035 (2006).
Sidi, S. et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2 and caspase-3. Cell. 133, 864–877, 10.1016/j.cell.2008.03.037 (2008).
Walters, K. B. et al. Muscle degeneration and leukocyte infiltration caused by mutation of zebrafish Fad24. Dev Dyn. 238, 86–99, 10.1002/dvdy.21821 (2009).
Rhodes, J. et al. Emi1 maintains genomic integrity during zebrafish embryogenesis and cooperates with p53 in tumor suppression. Mol Cell Biol. 29, 5911–5922 (2009).
Amin, R. H. et al. Biallelic, ubiquitous transcription from the distal germline Ig{kappa} locus promoter during B cell development. Proc Natl Acad Sci U S A. 106, 522–527, 10.1073/pnas.0808895106 (2009).
Villone, D. et al. Supramolecular interactions in the dermo-epidermal junction zone: anchoring fibril-collagen VII tightly binds to banded collagen fibrils. J Biol Chem. 283, 24506–24513, 10.1074/jbc.M802415200 (2008).
Robu, M. E., Zhang, Y. & Rhodes, J. Rereplication in emi1-deficient zebrafish embryos occurs through a Cdh1-mediated pathway. PloS one. 7, e47658, 10.1371/journal.pone.0047658 (2012).
Acknowledgements
This work was supported in part by the V Foundation (JR: 3263001) and PA Cure Funds (JR: 6913801). We thank FCCC Drs. Wiest, Balachandran and Seeger for feedback, Drs. Hardy and Li for real time PCR assistance, Dr. Eric Ross and Tianyu Li in the FCCC Bioinformatics and Biostatistics Facility, Alison Bilbee and Bruce Young in the Zebrafish Facility and the FCCC LAF, sequencing and cell sorting facilities.
Author information
Authors and Affiliations
Contributions
Contribution: J.R., W.P., M.D. and A.M.U. planned and performed experiments. J.R. and W.P. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Info
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Pimtong, W., Datta, M., Ulrich, A. et al. Drl.3 governs primitive hematopoiesis in zebrafish. Sci Rep 4, 5791 (2014). https://doi.org/10.1038/srep05791
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05791
This article is cited by
-
Unsupervised generative and graph representation learning for modelling cell differentiation
Scientific Reports (2020)
-
Effects of α-mangostin on embryonic development and liver development in zebrafish
Molecular & Cellular Toxicology (2020)
-
A conserved regulatory program initiates lateral plate mesoderm emergence across chordates
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.