« Prev Next »
Once it was determined that messenger RNA (mRNA) serves as a copy of chromosomal DNA and specifies the sequence of amino acids in proteins, the question of how this process is actually carried out naturally followed. It had long been known that only 20 amino acids occur in naturally derived proteins. It was also known that there are only four nucleotides in mRNA: adenine (A), uracil (U), guanine (G), and cytosine (C). Thus, 20 amino acids are coded by only four unique bases in mRNA, but just how is this coding achieved?
The Codon
The discordance between the number of nucleic acid bases and the number of amino acids immediately eliminates the possibility of a code of one base per amino acid. In fact, even two nucleotides per amino acid (a doublet code) could not account for 20 amino acids (with four bases and a doublet code, there would only be 16 possible combinations [42 = 16]). Thus, the smallest combination of four bases that could encode all 20 amino acids would be a triplet code. However, a triplet code produces 64 (43 = 64) possible combinations, or codons. Thus, a triplet code introduces the problem of there being more than three times the number of codons than amino acids. Either these "extra" codons produce redundancy, with multiple codons encoding the same amino acid, or there must instead be numerous dead-end codons that are not linked to any amino acid.
Preliminary evidence indicating that the genetic code was indeed a triplet code came from an experiment by Francis Crick and Sydney Brenner (1961). This experiment examined the effect of frameshift mutations on protein synthesis. Frameshift mutations are much more disruptive to the genetic code than simple base substitutions, because they involve a base insertion or deletion, thus changing the number of bases and their positions in a gene. For example, the mutagen proflavine causes frameshift mutations by inserting itself between DNA bases. The presence of proflavine in a DNA molecule thus interferes with the molecule's replication such that the resultant DNA copy has a base inserted or deleted.
Crick and Brenner showed that proflavine-mutated bacteriophages (viruses that infect bacteria) with single-base insertion or deletion mutations did not produce functional copies of the protein encoded by the mutated gene. The production of defective proteins under these circumstances can be attributed to misdirected translation. Mutant proteins with two- or four-nucleotide insertions or deletions were also nonfunctional. However, some mutant strains became functional again when they accumulated a total of three extra nucleotides or when they were missing three nucleotides. This rescue effect provided compelling evidence that the genetic code for one amino acid is indeed a three-base, or triplet, code.
Decoding the Genetic Code
However, at the time when this decoding project was conducted, researchers did not yet have the benefit of modern sequencing techniques. To circumvent this challenge, Marshall W. Nirenberg and Heinrich J. Matthaei (1962) made their own simple, artificial mRNA and identified the polypeptide product that was encoded by it. To do this, they used the enzyme polynucleotide phosphorylase, which randomly joins together any RNA nucleotides that it finds. Nirenberg and Matthaei began with the simplest codes possible. Specifically, they added polynucleotide phosphorylase to a solution of pure uracil (U), such that the enzyme would generate RNA molecules consisting entirely of a sequence of U's; these molecules were known as poly(U) RNAs. Each poly(U) RNA thus contained a pure series of UUU codons, assuming a triplet code. These poly(U) RNAs were added to 20 tubes containing components for protein synthesis (ribosomes, activating enzymes, tRNAs, and other factors). Each tube contained one of the 20 amino acids, which were radioactively labeled. Of the 20 tubes, 19 failed to yield a radioactive polypeptide product. Only one tube, the one that had been loaded with the labeled amino acid phenylalanine, yielded a product. Nirenberg and Matthaei had therefore found that the UUU codon could be translated into the amino acid phenylalanine. Similar experiments using poly(C) and poly(A) RNAs showed that proline was encoded by the CCC codon, and lysine by the AAA codon.
For example, when A and C were mixed with polynucleotide phosphorylase, the resulting RNA molecules contained eight different triplet codons: AAA, AAC, ACC, ACA, CAA, CCA, CAC, and CCC. These eight random poly(AC) RNAs produced proteins containing only six amino acids: asparagine, glutamine, histidine, lysine, proline, and threonine. Remember that previous experiments had already revealed that CCC and AAA code for proline and lysine, respectively. Thus, the four newly incorporated amino acids could only be encoded by AAC, ACC, ACA, CAA, CCA, and/or CAC. With the random sequence approach, the decoding endeavor was almost completed, but some work remained to be done.
Thus, in 1965, H. Gobind Khorana and his colleagues used another method to further crack the genetic code. These researchers had the insight to employ chemically synthesized RNA molecules of known repeating sequences rather than random sequences. For example, an artificial mRNA of alternating guanine and uracil nucleotides (GUGUGUGUGUGU) should be read in translation as two alternating codons, GUG and UGU, thus encoding a protein of two alternating amino acids. Translation of the artificial GUGU mRNA yielded a protein of alternating cysteine and valine residues. However, this technique alone could not determine whether GUG or UGU encoded cysteine, for example.
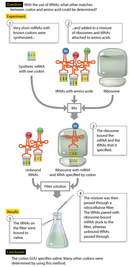
Next, Nirenberg and Philip Leder developed a technique using ribosome-bound transfer RNAs (tRNAs). They showed that a short mRNA sequence—even a single codon (three bases)—could still bind to a ribosome, even if this short sequence was incapable of directing protein synthesis. The ribosome-bound codon could then base pair with a particular tRNA that carried the amino acid specified by the codon (Figure 2).
Nirenberg and Leder thus synthesized many short mRNAs with known codons. They then added the mRNAs one by one to a mix of ribosomes and aminoacyl-tRNAs with one amino acid radioactively labeled. For each, they determined whether the aminoacyl-tRNA was bound to the short mRNA-like sequence and ribosome (the rest passed through the filter), providing conclusive demonstrations of the particular aminoacyl-tRNA that bound to each mRNA codon.
Degeneracy of the Amino Acid Code
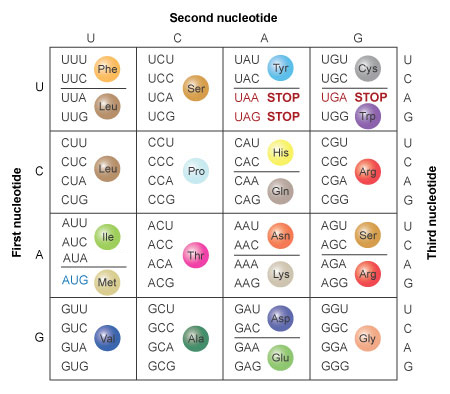
Examination of the full table of codons enables one to immediately determine whether the "extra" codons are associated with redundancy or dead-end codes (Figure 3). Note that both possibilities occur in the code. There are only a few instances in which one codon codes for one amino acid, such as the codon for tryptophan. Note also that the codon for the amino acid methionine (AUG) acts as the start signal for protein synthesis in an mRNA. Moreover, the genetic code also includes stop codons, which do not code for any amino acid. The stop codons serve as termination signals for translation. When a ribosome reaches a stop codon, translation stops, and the polypeptide is released.
References and Recommended Reading
Crick, F. H., et al. General nature of the genetic code for proteins. Nature 192, 1227–1232 (1961) (link to article)
Jones, D. S., Nishimura, S., & Khorana, H. G. Further syntheses, in vitro, of copolypeptides containing two amino acids in alternating sequence dependent upon DNA-like polymers containing two nucleotides in alternating sequence. Journal of Molecular Biology 16, 454–472 (1966)
Leder, P., et al. Cell-free peptide synthesis dependent upon synthetic oligodeoxynucleotides. Proceedings of the National Academy of Sciences 50, 1135–1143 (1963)
Nirenberg, M. W., Matthaei, J. H., & Jones, O. W. An intermediate in the biosynthesis of polyphenylalanine directed by synthetic template RNA. Proceedings of the National Academy of Sciences 48, 104–109 (1962)
Nirenberg, M. W., et al. Approximation of genetic code via cell-free protein synthesis directed by template RNA. Federation Proceedings 22, 55–61 (1963)
Nishimura, S., Jones, D. S., & Khorana, H. G. The in vitro synthesis of a co-polypeptide containing two amino acids in alternating sequence dependent upon a DNA-like polymer containing two nucleotides in alternating sequence. Journal of Molecular Biology 13, 302–324 (1965)



 Figure 1
Figure 1