« Prev Next »
Of course, these patterns of gene interaction apply not just to "normal" human traits, but to diseases as well. Many human diseases are multigenic, resulting from mutations in multiple genes that all affect a single phenotypic trait. Although the multifactorial nature of such diseases makes identification of the related genes difficult, researchers' awareness of this challenge has led to changes in the way diseases are studied.
Two Classes of Traits: Monogenic and Multigenic
For years, people have known that a number of traits result from mutations in single genes. For example, many cases of cystic fibrosis can be traced to a mutation in one particular gene, known as the CFTR gene. Monogenic ("single gene") diseases are usually as rare as they are severe, however, because selective pressure helps reduce the prevalence of deleterious mutations. This pressure is fast and effective in eliminating dominant mutations that act before sexual maturity. Nonetheless, a variety of monogenic diseases continue to persist in the human population. Some of these conditions are related to dominant mutations that either act after sexual maturity (e.g., Huntington's disease) or arise anew in individuals without being inherited. Still other monogenic diseases persist because they are caused by recessive mutations. Because selective pressure acts on only individuals who are homozygous for such alleles, a reservoir of these defective genes remains in the population.
In contrast to monogenic diseases, multigenic diseases result from less severe mutations in more than one gene. Any of these mutations alone might not affect a trait, but together, they can lead to significant phenotypic differences. Interaction among the phenotypic effects of different genes, often called epistasis, thus adds another layer of complexity to the study of disease inheritance. While epistasis dramatically complicates the task of determining the genetic underpinnings of human traits, recognizing that this phenomenon exists has changed the way in which disease is conceptualized and studied.
Traditional Approaches to the Study of Heritable Disease
To fully appreciate how multigenic inheritance complicates the study of disease, it's first important to understand some of the customary methods by which scientists have researched heritable conditions. Traditional approaches to studying such traits relied on the construction of pedigrees, or family trees, in which the trait in question was mapped onto family members, providing evidence for a pattern of inheritance. Later, with the dawn of the molecular age, strategies such as linkage mapping became popular. With linkage mapping, landmarks along the human genome, called single nucleotide polymorphisms (SNPs), are used as points of reference to determine what stretches of DNA are co-inherited with a certain trait or disease. Regions that are co-inherited are said to be in linkage disequilibrium; such regions are generally very close to one another and often physically linked (Figure 1; Kruglyak, 2008). Linkage mapping has been a valuable strategy in the study of several major human genes, including genes for type I diabetes.
While linkage mapping and related approaches are useful for identifying some genes, they have limitations. For instance, these methods generally aren't successful when studying traits that exhibit low heritability, traits that are highly influenced by environmental factors, or traits that are limited to groups of individuals with a small family tree. Furthermore, the variable features of particular traits can confuse researchers who are attempting to track the inheritance of these conditions. For example, some complex human diseases involve multiple symptoms that may fail to transmit in a single package; without the presence of a consistent set of symptoms among affected individuals, a disease can be difficult—if not impossible—to trace, even among members of a single family. Similarly, not all members of a family may be exposed to the same set of environmental factors (e.g., cigarette smoke or groundwater contaminants), thereby complicating the task of determining whether particular diseases are more strongly influenced by genetics or by surroundings.
New Strategies for Uncovering the Basis of Multigenic Diseases
Newer approaches to identifying disease loci adopt an unbiased, whole-genome approach. In other words, these methods compare the entire genome sequences of affected individuals to those of unaffected individuals, and any differences are noted as potential "hot spots" for the genes specifying a trait. In the past, this method was not pursued because the cost was prohibitive. However, recent advances in DNA sequencing technology, along with coordinated efforts to produce more complete, comprehensive lists of useful SNPs, have made this strategy realistic. In fact, whole-genome association studies have been used to study a growing list of human traits, including asthma (Moffatt et al., 2007) and restless leg syndrome (RLS) (Stefansson et al., 2007; Winkelmann et al., 2007).
What is especially interesting is that scientists are discovering unexpected genes associated with some of these complex disorders. Consider the case of RLS, a disease that causes sufferers to have pain and discomfort in their legs, particularly in the evening, thus disturbing their sleep. Although family histories have long suggested a genetic basis for RLS, scientists were not able to identify strong evidence supporting a genetic link until recently. Because RLS diagnosis is often found to coincide with iron deficiency in patients and symptoms are often alleviated by L-DOPA and other dopaminergic agonists, scientists expected that genes involved in either the iron metabolism pathway or the dopamine regulation pathway were also involved in RLS. However, investigation of genomic-level changes between RLS sufferers and healthy adults revealed several unexpected candidate genes, including polymorphisms in the Hox-family transcription factor gene MEIS1 (Figure 2; Winkelmann et al., 2007).
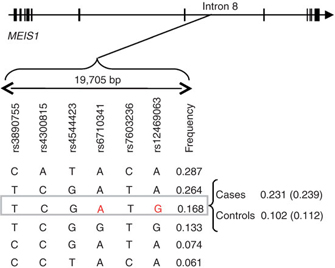
Interestingly, MEIS1 is known to play a role in specifying spinal motor neuron connectivity (Dasen et al., 2005), suggesting a tantalizing explanation for the neurological defects underlying RLS. Expression of MEIS1 is particularly strong in dopaminergic neurons and a region of the brain called the substantia nigra, which has been previously linked to RLS. This finding is consistent with the possibility that the MEIS1 transcription factor impacts expression of downstream target genes in these specific cell types, thereby leading to RLS. It is likely that other mechanisms can also impact the expression of RLS, given earlier genetic findings and the effects of iron deficiency. But consistent with the importance of MEIS1, spinal hyperexcitability is a known feature of RLS (Bara-Jimenez et al., 2000). As this example illustrates, genetic information provided by whole-genome association studies can inform efforts to understand the molecular bases of diseases—even those diseases with genetic regulatory interactions as part of their bases.
Complexities of Studying Human Genetic Disorders
As the truly complex nature of the genetics of human disease becomes increasingly apparent, it is important to remember that additional factors can complicate matters even further. For example, environmental effects can also impact interactions among genes, as toxins present in the environment may further dampen already weakened pathways. However, knowledge of environmental impact can provide two important benefits. First, this knowledge can be used to create better and more accurate pedigrees, wherein information about patient environments is incorporated into the pedigree. Second, knowledge of the impact of environmental factors on multigenic diseases can lead to improved, highly personalized prevention programs for individuals who carry the related genes, thereby informing health care choices for individuals concerned with preventing the onset of certain diseases. Although detailed knowledge of patient pedigrees has raised some ethical concerns in the past, the recent passage of the Genetic Information Nondiscriminatory Act (GINA) by Congress should make efforts to study genetic and environmental influences on disease even more tangible.
References and Recommended Reading
Dasen, J. S., et al. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell 123, 477–491 (2005)
Kingsmore, S. F., et al. Genome-wide association studies: Progress and potential for drug discovery and development. Nature Reviews Drug Discovery 7, 221–230 (2008) doi:10.1038/nrd2519 (link to article)
Kruglyak, L. The road to genome-wide association studies. Nature Reviews Genetics 9, 314–318 (2008) doi:10.1038/nrg2316 (link to article)
Moffatt, M. F., et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448, 470–473 (2007) (link to article)
Nisticò, L., et al. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Human Molecular Genetics 5, 1075–1080 (1996)
Stefansson, H., et al. Neuregulin 1 and susceptibility to schizophrenia. American Journal of Human Genetics 71, 877–892 (2002)
---. A genetic risk factor for periodic limb movements in sleep. New England Journal of Medicine 357, 639–647 (2007)
Winkelmann, J., et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nature Genetics 39, 1000–1006 (2007) (link to article)



 Figure 1
Figure 1


























