Abstract
Myxoid/round cell (RC) liposarcomas (MLS) were originally classified into two distinct populations based on histological differences; a myxoid component and a RC component. It is notable that, depending on an increase of the RC component, the prognosis significantly differs. Hence, the RC component is associated with metastasis and poor prognosis. However, the molecular mechanisms that contribute to the malignancy of the RC component still remain largely unknown. Here, we report microRNA-135b (miR-135b), a key regulator of the malignancy, highly expressed in the RC component and promoting MLS cell invasion in vitro and metastasis in vivo through the direct suppression of thrombospondin 2 (THBS2). Decreased THBS2 expression by miR-135b increases the total amount of matrix metalloproteinase 2 (MMP2) and influences cellular density and an extracellular matrix structure, thereby resulting in morphological change in tumor. The expression levels of miR-135b and THBS2 significantly correlated with a poor prognosis in MLS patients. Overall, our study reveals that the miR-135b/THBS2/MMP2 axis is tightly related to MLS pathophysiology and has an important clinical implication. This work provides noteworthy evidence for overcoming metastasis and improving patient outcomes, and sheds light on miR-135b and THBS2 as novel molecular targets for diagnosis and therapy in MLS.
Similar content being viewed by others
Introduction
Liposarcoma (LPS) is one of the most common soft tissue sarcomas in adults, and it is classified into three subtypes according to histopathological examination: well-differentiated/dedifferentiated LPS, myxoid/round cell (RC) LPS (MLS) and pleomorphic LPS. MLS accounts for 30–35% of all LPS cases and approximately 10–15% of all adult soft tissue sarcomas.1 MLS covers a wide morphological spectrum of tumors ranging from pure myxoid tumors to hypercellular RC tumors. MLS with a more than 5% RC component is associated with poor prognosis and metastasis,2, 3 and patients with this type of MLS typically receive combined chemotherapy and radiotherapy. The 5-year survival rate is significantly lower for MLS patients with a more than 5% RC component (40–60%) compared with MLS patients with a less than 5% RC component (70–90%).4, 5, 6 The characterization of the RC component using hematoxylin and eosin staining and the RC ratio remains the gold standard for predicting MLS prognosis. However, it can be difficult to determine an accurate percentage of the RC component because of sampling bias, especially with small biopsy specimens,7 and ambiguous boundaries.8 Therefore, further investigations into the mechanisms that contribute to the malignancy of the RC component and the identification of new diagnostic markers are required to improve the reliability in predicting MLS patient outcomes.
microRNAs (miRNAs) are small non-coding RNAs of approximately 22 nucleotides that are related to various biological processes such as development, differentiation, apoptosis and proliferation.9 The abnormal expression of miRNAs has been observed in solid and hematopoietic tumors, resulting in tumor initiation and progression.9, 10 In MLS, suppression of miR-486 by TLS-CHOP, a fusion transcript resulting from the 12q13 and 16p11 translocations in <95% of MLS patients, increases the expression of plasminogen activator inhibitor-1, thereby enhancing cell growth.11 In a copy number variation study, miR-26a-2 has been found to be amplified and highly expressed in 22 MLS patients and affects cell proliferation and survival by inhibiting a regulator of chromosome condensation and BTB domain containing protein 1.12 Global miRNA expression analyses using microarrays from high-grade soft tissue sarcomas13 and various subtypes of LPS14 have enabled the identification and classification of MLS tumors based on their miRNA expression profiles.
Despite evidence of miRNA deregulation in MLS, little is known regarding the contribution of miRNAs in the RC component malignancy and the molecular and clinical implications of miRNAs in MLS. Here, we report that miR-135b is highly expressed in the RC component and promotes invasion in vitro and metastasis in vivo. Moreover, miR-135b triggers histopathological abnormalities via an enhanced degradation of the extracellular matrix by the increased matrix metalloproteinase 2 (MMP2) through the direct downregulation of thrombospondin 2 (THBS2). Finally, we show statistical significance and clinical relevance of miR-135b and THBS2, which correlated with a poor prognosis in MLS. Our findings demonstrate a critical pathway involving miR-135b and THBS2 that could represent a valuable prognostic biomarker and therapeutic target for MLS patients.
Results
miR-135b overexpression in the RC component is associated with MLS cell invasion in vitro
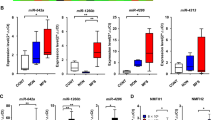
To identify the miRNAs responsible for malignancy of the RC component in MLS, we performed laser-capture microdissection-based miRNA microarray analyses of the myxoid and RC components (Figure 1a). Eight formalin-fixed, paraffin-embedded (FFPE) samples from seven MLS patients who never received chemotherapy or radiotherapy (six primary tissues and one recurrent tissue) contained sections with a more than 5% RC component. On the basis of the array analysis, 16 candidate miRNAs were identified as exhibiting significantly high expression in the RC component (Figure 1b). Eleven out of 16 miRNAs were validated by qRT–PCR, and we focused on miR-135b because it displayed the highest upregulation in the RC component (Figure 1c and Supplementary Figure 1). miR-135b expression was significantly higher in three MLS cell lines compared with adipose tissue-derived mesenchymal stem cells (Figure 1d) and also higher in MLS patient tumor tissues (regardless of the cellular component, n=30) compared with adjacent normal tissues (n=25) (Figure 1e). Notably, the increased miR-135b levels correlated with the RC ratio (Figure 1f). As the RC component is considered as a key factor in cancer invasion, we tested miR-135b effect in cell invasiveness in vitro. First, we performed a serial cell invasion assay using a Matrigel invasion chamber (Figure 1g) and obtained highly invasive clones (Supplementary Figure 2a), which overexpressed miR-135b compared with parental cells (Figure 1g). Next, we transiently transfected the 2645-94 and 402-91 cell lines with the miR-135b mimics. It significantly promoted the invasive ability of both MLS cell lines (Figure 1h and Supplementary Figures 2b-d). However, increased miR-135b expression did not affect cell growth (Supplementary Figure 2e) or migratory activity (data not shown). These results suggest that miR-135b is an oncogenic miRNA highly expressed in the RC component and has a critical role in MLS cell invasion.
miR-135b is highly expressed in the RC component and is associated with MLS invasiveness. (a) Collection of tissue fragments from the myxoid and RC components of clinical FFPE samples with laser-capture microdissection. (b) Heat map of miRNA microarray analysis represents the significantly different expression levels of miRNAs between the two groups. The color value indicates log2 (normalized data). (c) qRT–PCR analysis showing the 16 miRNA expression levels chosen from the results of microarray. The levels are expressed as the fold change of the RC component relative to the myxoid component. (d) qRT–PCR analysis showing the expression levels of miR-135b in human MLS cell lines (2645-94, 402-91 and 1955-91) relative to adipose tissue-derived mesenchymal stem cells as a control; n=4 per group. (e and f) qRT–PCR analysis showing the expression levels of miR-135b in MLS tumors and adjacent normal tissues. (g) Schematics of invasive cell selection (left upper panel). qRT–PCR analysis showing the expression levels of miR-135b in 2645-94 parental and invasive cells (left lower panel). Images of invasion assay after enrichment selections (right panels). Scale bars, 300 μm. (h) Quantification (left panel) and images of the effect on invasion of 2645-94 cells after transfection (right panels). The invasive values were normalized to those of the control cells; n=4 per group. Scale bars, 200 μm. Student’s t test was used for statistical analyses. All data are presented as the mean±s.e.m. *P<0.1; **P<0.05; ***P<0.01.
THBS2 is a direct target of miR-135b in MLS
To identify the target of miR-135b, we integrated expression data of genes and miRNA with in silico approaches (five target prediction algorithms). Four genes were isolated as miR-135b targets on satisfying three of the following conditions (Figure 2a). Genes that were (i) downregulated in the RC component by 1.5-fold compared with the myxoid component in the mRNA sequencing of the total RNA derived from the clinical FFPE samples; (ii) downregulated in miR-135b-overexpressing cells by fivefold compared with the negative control (NC) in the mRNA microarray of the 2645-94 cells; and (iii) predicted as targets of miR-135b using in silico analysis. THBS2, interleukin 1 receptor 1 (IL1R1), glutamate receptor, ionotropic, AMPA 3 (GRIA3), and collagen, type V, alpha 1 (COL5A1) were selected as final candidate targets for miR-135b. To test them, miR-135b was transiently overexpressed in MLS cells, and the expression levels of the four genes were quantified by qRT–PCR. Only THBS2 and IL1R1 expressions were significantly decreased in miR-135b-overexpressing cells (Figure 2b). The expression level of THBS2 in all three MLS cell lines was significantly lower than in adipose tissue-derived mesenchymal stem cells, but that of IL1R1 was not (Figure 2c and Supplementary Figure 3a). Protein level of THBS2 in all three MLS cell lines was lower than in adipose tissue-derived mesenchymal stem cells (Supplementary Figure 3b) and also clearly decreased after transient transfection with the miR-135b mimics (Figure 2d and Supplementary Figure 3c). To further verify whether THBS2 is a direct target of miR-135b, we performed a 3′ UTR assay. The luciferase activity level was significantly reduced by approximately 40% in the cells cotransfected with the miR-135b mimics compared with the NC. In contrast, the cotransfection with the mutated form of 3′ UTR resulted in no significant change in luciferase activity (Figure 2e). Taken together, our data strongly indicate that THBS2 is a direct target of miR-135b in MLS.
THBS2 is a direct target of miR-1z35b in MLS. (a) Venn diagram of the candidate target mRNAs of miR-135b based on the mRNA sequence of clinical FFPE samples, the mRNA microarray of 2645-94 cells after transient transfection of the miR-135b mimics and the NC, and in silico analysis. (b) qRT–PCR analysis showing the expression levels of four candidate mRNAs in transfected 2645-94 cells by the miR-135b mimics and the NC; n=5 per group. (c) qRT–PCR analysis showing the expression levels of THBS2 in human MLS cell lines (2645-94, 402-91 and 1955-91) compared with adipose tissue-derived mesenchymal stem cells as a control; n=3 per group. (d) Western blot analysis of THBS2 after transient transfections with the miR-135b mimics and the control in 2645-94. (e) Schematics of the miR-135b-binding site within the 3′ UTR of THBS2 and its mutated sequence (left panel). Luciferase activities (right panel) verifying that the suppression of THBS2 by miR-135b was compared with control measured in 2645-94; n=3 per group. Student’s t test (b, e) and Dunnett's test (c) were used for statistical analyses. All data are presented as the mean±s.e.m. **P<0.01; ***P<0.001.
Inverse correlation between miR-135b and THBS2 in MLS
We next looked at the expression levels of THBS2 in clinical samples of MLS by qRT–PCR with the FFPE samples of the myxoid and RC components. We found that THBS2 expression was lower in the RC component compared with the myxoid component in six of the eight samples (Supplementary Figure 4a). Likewise, a decreased expression level of THBS2 was confirmed by immunohistochemistry in the RC component compared with the myxoid component in the clinical MLS FFPE samples (Figure 3a). It was further confirmed in a tissue microarray containing nine myxoid LPS tissues (positivity: six out of nine, 67%) and three RC LPS tissues (positivity: one out of three, 33%) (Figure 3b and Supplementary Figure 4b). We then scrutinized the expression levels of THBS2 in MLS patient tumors. THBS2 expression was significantly decreased in tumors (Figure 3c) and tended to be inversely correlated with miR-135b depending on the RC ratio (Figure 3d). In comparison between MLS tumors and adjacent normal tissue, THBS2 clearly showed inverse expression patterns to miR-135b (Figure 3e). To investigate the functional effect of THBS2 on the invasiveness of MLS cells, we knocked down THBS2 by three siRNAs with different sequences (Supplementary Figures 5a and b and Supplementary Figures 6a and b). As a result, the silencing of THBS2 significantly enhanced the invasion ability of MLS cells (Figure 3f, Supplementary Figures 5c and d and Supplementary Figure 6c). Cell proliferation analysis suggested that the decreased expression of THBS2 did not affect cell growth (Supplementary Figure 5e and Supplementary Figure 6d). These findings showed that miR-135b blocked THBS2 expression in the RC component and miR-135b/THBS2 axis contributes to the invasiveness of MLS cells.
Inverse correlation between miR-135b and THBS2 in MLS. (a and b) Representative case and section of THBS2 immunohistochemical staining of clinical FFPE and tissue microarray with MLS tissue sections. Scale bars, 100 μm. (c) qRT–PCR analysis showing the expression levels of THBS2 in MLS tumors and adjacent normal tissues. (d) qRT–PCR analysis showing the expression levels of THBS2 in MLS tumors based on the ratio of the RC component. (e) Scatter plots showing the expression levels of miR-135b and THBS2 in MLS tumors compared with adjacent normal tissues. (f) Quantification (left panel) and images (right panel) of the effect of THBS2 silencing by the siRNAs on the invasion in 2645-94 cells. The invasion values were normalized to those of the control; n=4 per group. Student's t test was used for statistical analyses. All data are presented as the mean±s.e.m. *P<0.05; ***P<0.01.
miR-135b increases the total amount of MMP2 in extracellular environments by repressing THBS2 expression
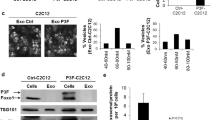
To confirm the association between miR-135b and MLS progression, we performed a pathway analysis from the mRNA sequencing comparing the myxoid and RC components. Ten pathways (nine upregulated and one downregulated in the RC component) were identified as significant with an FC<−2.0 (Figure 4a). We looked into the pathway of matrix metalloproteinases, which are involved in invasion and metastasis. We focused on the MMP2 of them all, one of the genes in the pathway ‘matrix metalloproteinases’ because previous studies have demonstrated the interaction between THBS2 and MMP2.15, 16, 17 Therefore, we first determined the expression levels of MMP2 in clinical frozen tissue samples. MMP2 expression was significantly higher in MLS tumors than in adjacent normal tissues in frozen tissue specimens (Figure 4b and Supplementary Figure 7a) and also exhibited the higher protein existence in the RC component than in the myxoid component (Supplementary Figure 7b). So, we conducted a gelatin zymography assay with the cell conditioned media as described in Figure 4c, to evaluate the existence of pro- and active-MMP2 after transient transfection with the miR-135b mimics, the THBS2 siRNAs and the NC. When treated with the conditioned media derived from the miR-135b mimic-transfected cells, the amount of both pro- and active-MMP2 was enhanced as well as the THBS2 siRNAs (Figure 4d and Supplementary Figure 8). Collectively, our data demonstrate that miR-135b induced the accumulation of total MMP2 through the downregulation THBS2, leading to increased invasiveness in MLS.
miR-135b increases the activity of MMP2 by repressing THBS2 expression. (a) Pathway analysis comparing the myxoid and RC components in clinical FFPE samples. The pathways with a significant difference above twofolds are listed (red bar: RC>myxoid, blue bar: RC<myxoid). (b) qRT–PCR analysis showing the expression levels of MMP2 in MLS tumors and adjacent normal tissues. (c) Schematics of sample preparation procedure for gelatin zymography. (d) Gelatin zymography of proteins secreted into the conditioned media after treatment with the miR-135b mimics, the THBS2 siRNAs and the NC (upper panel) and its quantification of band intensity of pro-MMP2 (left lower panel) and active-MMP2 (right lower panel); n=4 per group. Permutation test (a) and Student’s t test (b, d) were used for statistical analyses. All data are presented as the mean±s.e.m. *P<0.05, **P<0.01.
miR-135b enhances tumor enlargement and triggers histopathological abnormality in tumors in vivo
To examine the effects of miR-135b on tumor enlargement and histopathological changes in vivo, we set up a MLS xenograft model using intramuscular injections into NOG mice. For tumor growth monitoring, we established 2645-94 cell lines stably expressing miR-135b or control miRNA with the pLV-GFP-Luc plasmid DNA as described in the Materials and methods section. We confirmed the increased expression of miR-135b (Supplementary Figure 9a) and the decreased expression of THBS2 at mRNA levels (Supplementary Figure 9b) in the established 2645-94 GFP-Luc-miR-135b cells compared with the control cells. We also confirmed that these cells exhibited increased invasiveness in vitro (Supplementary Figure 9c). Ten weeks after transplantation, five mice from each group were killed, and their primary tumors were extracted for histopathological examination. On the basis of bioluminescence imaging, the 2645-94 GFP-Luc-miR-135b cells showed enhanced tumor enlargement compared with that of control cells (Figures 5a and b). Interestingly, there are significant increases of hypercellular area in morphology in miR-135b-overexpressing tumors relative to in control tumors (Figure 5c and Supplementary Figure 9d). Tumors derived from miR-135b-overexpressing cells had increased cellular densities. We observed a decreased expression of THBS2 in tumors derived from miR-135b-overexpressing cells by immunohistochemistry for THBS2 (Figure 5d) and reduced extracellular collagen matrix by Masson’s trichrome staining (Figure 5e), as compared with control tumors. These results suggested that miR-135b enhanced the expansion of MLS cells and induced histopathological alterations in vivo.
miR-135b enhances tumor enlargements and triggers histopathological abnormalities of MLS tumors in vivo. (a) Bioluminescence images of primary tumor growth in each group 10 weeks after transplantation (five mice per group). (b) Extracted tumors (left panel) and quantification of tumor volume (right panel) at primary site of each group. (c) Whole microscopic images of hematoxylin and eosin (H&E) staining and overlaid images, with three types of contents shown in different colors, of primary tumor derived from each group (left panel). The hypercellular areas are shown in purple, hypocellular areas in light blue and mouse muscle in red. The quantification of the percentage of hypercellular area per whole tumor (right upper panel) and the distribution of cellular density (right lower panel); n=5 per group. (d) Representative images of H&E staining and immunohistochemical staining for THBS2 of each group. (e) Representative images of Masson’s trichrome staining of each group. Student’s t test (b, c (right upper panel)) and Fisher's exact test (c (right lower panel)) were used for statistical analyses. All data are presented as the mean±s.e.m. Scale bar, 300 μm (c), 50 μm (d, e). *P<0.05.
Silencing of miR-135b inhibits the development of lung metastasis in established high lung-metastatic MLS cells in vivo
To evaluate the effect of miR-135b on metastasis in vivo, we established high lung-metastatic 2645-94 cells, named 2645-94 GFP-Luc LM2 (LM2) (Figure 6a). These cells were characterized by rapid and high lung metastasis frequencies (Table 1) and increased tumorigenic ability in vivo (Supplementary Figures 10a and b). Moreover, these cells, compared with 2645-94 GFP-Luc (parental) cells, also displayed increased invasion and migratory capacities in vitro (Supplementary Figures 10c and d). To confirm that the LM2 cells originated from MLS cells, we detected the TLS-CHOP fusion gene by nested PCR (Supplementary Figure 10e). The expression level of miR-135b was significantly higher in tumors derived from LM2 cells than from parental cells (Figure 6b). We also validated the decreased expression level of THBS2 in tumors derived from LM2 cells relative to the expression in parental cells by immunohistochemistry for THBS2 (Supplementary Figure 10f). We made stable lines of miR-135b knockdown (KD) and the control 2645-94 GFP-Luc LM2 cells. We checked the decreased expression levels of miR-135b (Supplementary Figure 11a) and the increased expression levels of THBS2 (Figure 6c), and attenuated invasiveness in vitro (Figure 6d) in the established 2645-94 GFP-Luc LM2 miR-135b-KD cells. Three weeks after intramuscular transplantation of LM2 cells, 10 mice from each group were killed, and their lungs were extracted for the evaluation of lung metastases. On the basis of bioluminescence imaging, lung metastases were observed in four 2645-94 GFP-Luc LM2 miR-135b-KD cell-transplanted mice and in seven mice transplanted with control cells (Figure 6e). A histological analysis showed that the miR-135b-KD reduced the pulmonary metastases of this MLS cell line (Figure 6f) and induced the significant resumption of THBS2 expression in the metastatic foci (Supplementary Figure 11b). These findings indicated that miR-135b contributed to metastasis in vivo.
Silencing of miR-135b in established high lung-metastatic MLS cells reduces lung metastasis in vivo. (a) Schematics of the establishment of 2645-94 GFP-Luc high lung-metastatic cells, termed LM2 cells, using NOG mice. (b) qRT–PCR analysis showing the expression levels of miR-135b in tumors derived from 2645-94 GFP-Luc parental cells and LM2 cells; n=5 per group. (c) qRT–PCR analysis of the expression levels of THBS2 in miR-135b stable knockdown cells and control cells of 2645-94 GFP-Luc LM2; n=4 per group. (d) Quantification (left panel) and images (right panels) of the effect on invasion of 2645-94 GFP-Luc LM2 cells after stable miR-135b knockdown. The invasive values were normalized to those of the control cells; n=4 per group. Scale bars, 200 μm. (e) Bioluminescence images of the chest regions in each group 3 weeks after transplantation (10 mice per group). Table showing the number of lung metastasis in 2645-64 LM2 cells. LM, lung metastasis. (f) Microscopic pictures of hematoxylin and eosin staining of the extracted lung (left and middle panels), the number of metastatic foci per mouse (right upper panel) and the distribution of metastatic focus size (right lower panel). The black arrowhead represents metastatic nodule in lung; n=10 per group. Scale bar, 1 mm (left) and 200 μm (middle). Student’s t test (b–d, and f (right upper panel)) and Fisher’s exact test (f (right lower panel)) were used for statistical analyses. All data are presented as the mean±s.e.m. *P<0.05; ***P<0.001.
Clinical relevance of high expression of miR-135b and low expression of THBS2 linking to poor prognosis in MLS
Given that the RC component in MLS is a risk factor associated with poor prognosis, miR-135b and THBS2 expression levels also could be critical factors to predict prognosis in MLS. To this end, we examined the survival outcomes of 64 MLS patients (Supplementary Table I). A Kaplan–Meier analysis revealed that the presence of a more than 5% RC component closely correlated with poor prognoses in MLS patients (log-rank test, P=0.001 for disease-free survival, Figure 7a). Next, we examined the correlation between MLS patient outcomes and the expression levels of miR-135b and THBS2 in clinical FFPE biopsy samples. Kaplan–Meier analyses indicated that high expression levels of miR-135b and low expression levels of THBS2 were significantly associated with poor prognoses (log-rank test, P=0.006 and 0.001 for disease-free survival, respectively, Figures 7b and c). Consequently, the miR-135b and THBS2 expression levels appeared to be more relevant than the ratio of the RC component in predicting clinical outcomes (log-rank test, P=0.016 for disease-free survival, Figure 7d). These data strongly suggest that miR-135b and THBS2 could be valuable prognostic biomarkers reflecting the clinical behaviors of MLS tumors.
The high expression of miR-135b and low expression of THBS2 correlates with poor survival in MLS patients. (a) Kaplan–Meier analysis of disease-free survival according to the RC component in 64 MLS patients. (b and c) Kaplan–Meier analysis of disease-free survival according to the expression levels of miR-135b and THBS2 in 19 MLS patient biopsy samples. Cutoff values were determined with the best Youden index. (d) Kaplan–Meier analysis of disease-free survival according to the RC component in 19 MLS patient biopsy samples. (e) Schematics of miR-135b/THBS2/MMP2 axis in MLS. miR-135b increases the total amount of MMP2 in extracellular matrix by the directly suppression of THBS2 and contributes to tumor progression and histopathological change in MLS. Log-rank tests were used for statistical analyses (a–d).
Discussion
In previous research, MLS cases were initially categorized as either myxoid LPS or RC LPS. However, because both myxoid and RC LPSs were observed on the same morphological continuum and contained the same the 12q13 and 16p11 translocations (resulting in the TLS-CHOP fusion transcript), the two types of sarcomas have been grouped together as myxoid and RC LPS.18 More than 95% of MLS patients have the TLS-CHOP fusion transcript; TLS-CHOP is a key molecule associated with tumor initiation and the inhibition of adipogenesis.19, 20 However, the RC component is also a risk factor associated with metastasis and poor prognosis. Thus, the presence of TLS-CHOP in MLS patients cannot fully account for the malignancy or the morphological spectrum. There are several reports concerning the relationship between the myxoid and RC components. Some groups have reported that the myxoid and RC components are the well-differentiated and the poorly differentiated MLS populations, respectively.21, 22 In other studies, further genetic damage,23 PI3K/Akt pathway activation24 and heparin-binding EGF-like growth factor secretion by tumor-associated macrophages25 following MLS initiation by TLS-CHOP were suggested as factors leading to the transformation from the myxoid to the RC component. However, many of the details regarding the relationship between the two components are unknown. We hypothesized that overexpressed miRNAs in the RC component may contribute to tumor progression and the histopathological differences from the myxoid component.
In this study, we found that miR-135b was more highly expressed in the RC component and was associated with cell invasion in vitro and metastasis in vivo. To the best of our knowledge, this is the first report to compare the miRNA profiles of the myxoid and RC components and to investigate in detail the effect of miRNA in MLS progression. Previous studies have reported that miR-135b is dysregulated in colorectal cancer,26, 27, 28, 29 lung cancer,30 pancreatic cancer,31 liver cancer32 and osteosarcoma,33 indicating that miR-135b generally functions as an oncogenic miRNA. Lin et al.30 have reported that miR-135b promotes tumor growth, cellular invasiveness and metastasis in lung cancer by repressing multiple key components of the Hippo pathway network with the tumor suppressor LZTS1. Li and colleagues32 have demonstrated that miR-135b promotes tumor invasion and metastasis by regulating RECK and EVI5 in hepatocellular carcinoma. In agreement with these reports, we found that miR-135b was involved in promoting MLS cell invasion and metastasis.
We further demonstrated that miR-135b increased the total amount of MMP2 in the extracellular matrix through the direct suppression of THBS2. Previous studies have reported that low expression levels of THBS2 are associated with poor prognoses in gastric cancer,34 prostate cancer,35 colon cancer36 and pancreatic cancer.37 Moreover, THBS2 contributes to cancer progression, such as invasion37 and metastasis.38 In the current study, patient sample no. 3 of FFPE displayed a high expression level of THBS2 in the RC component despite the high miR-135b expression. We considered that this contradictory expression between miR-135b and THBS2 might be due to polymorphisms, such as single nucleotide mutations and insertion/deletions, at miRNA target sites.39, 40, 41 However, we obtained consistent expression patterns between miR-135b and THBS2 in almost all clinical samples and our in vitro experiments strengthened the inverse correlation. Moreover, THBS2 binds to both pro-MMP2 and active-MMP2 with high affinity. Then, this complex is cleared from the pericellular environment by binding with the low-density lipoprotein receptor-related protein receptor and directed to the lysosomes for degradation. This mechanism has an important role in the regulation of extracellular MMP2 levels.15, 16, 17 Previous studies have reported that membrane type 1-MMP (MT1-MMP) is a key activator of MMP2.42 However, MT1-MMP expression did not change between the myxoid and RC components in our study. Consequently, our data suggest that a molecular mechanism independent of MT1-MMP based on the regulation of THBS2 expression by miR-135b enhances the accumulation of total MMP2 in the extracellular environment, leading to cell invasion and metastasis in MLS.
From the results of the animal experiments, we found that miR-135b overexpression triggered histopathological changes as evidenced by increased cellular densities and decreased extracellular collagen matrix. These histopathological findings were similar to the RC component observation in human MLS. Previous studies have also identified gene alterations between the myxoid and RC components using clinical tumors of MLS patients. Oda et al.43 have reported that a reduced protein expression of the cell cycle regulatory genes, p16INK and p14ARF, is more frequently observed in the RC component than in the myxoid component and may be one of the critical events during MLS progression. Furthermore, highly malignant MLS with RC components showed longer telomeres, stronger telomerase activity, and higher hTERT and c-MYC expression levels, compared with the pure myxoid variants.44 From these studies, cell growth ability may be associated with the difference between each pathological region. Consistently with previous reports, our study shows that cell cycle-related pathways were upregulated in the RC component (Figure 4a). However, the miR-135b overexpression did not affect cell growth in vitro (Supplementary Figure 2e). On the other hand, the stable miR-135b-overexpressing cells exhibited enhanced tumorigenic abilities in vivo. We propose that this discrepancy was caused by the tumor microenvironment, the effect of MMP2, and the nature of the 2645-94 cells. First, the tumor microenvironment, which includes cells such as macrophages and endothelial cells as well as extracellular matrix components, proteases and cytokines,45 has a critical role during tumor progression and metastasis. Second, a possible explanation for this discrepancy is that the increased MMP2 in the extracellular matrix as a result of THBS2 downregulation by miR-135b degrades the extracellular collagen matrixes; thus, tumor cells are likely to spread towards their surroundings. Third, the original 2645-94 cells have very weak malignancy, including tumorigenicity in vivo. These cells did not survive in nude mice (BALB/cAJcl-nu/nu), SCID mice (C.B-17/Icr-scid/scid Jcl) or NOD/SCID mice (NOD/ShiJic-scid Jcl) in our preliminary animal experiments. Therefore, it is possible that cell death is induced in some of the control cells, even if in our in vivo model using NOG mice, which may cause the differences in the tumor growth between the control and miR-135b-overexpressing cells. Our findings suggest that histopathologic differences between the myxoid and RC components may be due to the miR-135b/THBS2/MMP2 pathway. The increased total amount of MMP2 triggers the degradation of the extracellular matrix and promotes the invasion of tumor cells in the extracellular space (Figure 7e). We propose that this molecular mechanism, independent of cell proliferation or other mechanisms previously reported, may contribute to the morphological spectrum of MLS.
In addition, we indicated that high expression levels of miR-135b and low expression levels of THBS2 may relate to the good ability of the RC component to predict MLS patient outcomes (Figures 7b and c). Presently, no reliable biomarkers have been identified in sarcomas, including MLS. Although various mRNAs46 and miRNAs47 have been reported as disease-specific markers in sarcomas, no targeting molecules have been developed for clinical applications. Recently, it has been shown that extracellular miRNAs are contained in various bodily fluids and can circulate in the blood.48, 49 These cell-free miRNAs could be potential diagnostic and prognostic biomarkers for various types of cancer. The expression profiles of circulating miRNAs in serum and plasma are similar to those of miRNAs derived from tumor tissues.50, 51 Circulating miR-135b levels in the plasma of endometrial carcinoma patients have been observed to be upregulated and may serve as a novel and less invasive biomarker for diagnosis.52 Similarly, in stool samples, the level of miR-135b is significantly higher in patients with colorectal cancer or adenomas than in non-cancerous patients.53 There is a continued need to increase the number of available MLS tissue samples and to further investigate the expression levels of miR-135b and THBS2 in blood. Therefore, although more number of the available MLS tissue samples as well as patient-matched body fluidic samples such as blood are necessary to further investigate the possibility of miR-135b and THBS2 as biomarkers, they may be promising biomarkers reflecting the clinical outcome of MLS patients.
Materials and methods
Clinical samples
This study was allowed by the Institutional Review Board at the National Cancer Center (Number: 2013-102). All clinical information and materials were obtained after receiving informed consent from the patients. MLS tissue samples, including both FFPE and frozen tissue samples from MLS patients who did not receive chemotherapy or radiotherapy, were collected at the National Cancer Center Hospital of Japan between January 1997 and December 2015. For miRNA microarray analyses, we selected eight FFPE samples from MLS patients with more than 5% RC component according to pathological reports (Supplementary Table II). Furthermore, we collected 30 MLS tumors, 25 adjacent normal muscle tissue samples (Supplementary Tables III and VI) and 19 FFPE biopsy samples (Supplementary Table V) from MLS patients for quantitative RT–PCR (qRT–PCR), regardless of the RC component content. Diagnoses were based on standard histopathological criteria according to the WHO classification of Tumors of Soft Tissue and Bone.
Cell culture
Human MLS cell lines 2645-94, 1955-91 and 402-91 were previously established54, 55 and provided by Prof M Kuroda (Tokyo Medical University). The human adipose-derived stem cell line was purchased from Invitrogen (Carlsbad, CA, USA) (Lot. 1212). All cell lines were cultured in RPMI 1640 with 25 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer (Life Technologies, Carlsbad, CA, USA). All media were supplemented with 10% heat-inactivated FBS (Life Technologies) and antibiotic-antimycotics (Invitrogen). The cells were maintained under 5% CO2 in a humidified incubator at 37 °C.
miRNA and mRNA microarray analysis
To detect the miRNAs in the myxoid and RC components of MLS FFPE samples, 100 ng of total RNA was labeled using miRNA Complete Labeling and Hyb Kits (Agilent Technologies, Santa Clara, CA, USA) and hybridized on an Agilent SurePrint G3 Human miRNA 8x60K Rel.19 array (design ID: 046064) according to the manufacturer’s protocol. Hybridization signals were scanned using a DNA microarray scanner (Agilent Technologies), and the scanned images were quantified using Agilent Feature Extraction software. We applied a ⩾5-fold change in signal intensity to identify significant differences in gene expression. The raw and normalized microarray data are available in the Gene Expression Omnibus database (accession number GSE73448).
To detect the mRNAs in cells derived from 2645-94 cells transfected with miR-135b mimics or NC, 100 ng of total RNA was labeled using an Agilent Low Input Quick Amp Labeling Kit, one-color (Agilent Technologies) and hybridized using Agilent SurePrint G3 Human GE v2 8x60K Microarray (Design ID: 039494) according to the manufacturer’s instructions (Protocol for Use with Agilent GE Microarrays Version 6.7). Hybridization signals were scanned using a DNA microarray scanner (Agilent Technologies), and the scanned images were quantified using Agilent GeneSpring software. We applied a ⩾5-fold change in signal intensity to identify significant differences in gene expression. The raw and normalized microarray data are available in the Gene Expression Omnibus database (accession number GSE73347).
mRNA sequencing and pathway analysis
A total of 100 ng of total RNA obtained from each FFPE sample was subjected to a sequencing library construction using a TruSeq RNA Access library prep kit (Illumina, San Diego, CA USA) according to the manufacturer’s protocols. The quality of the libraries was assessed with an Agilent 2200 TapeStation High Sensitivity D1000 assay (Agilent Technologies). The pooled libraries were sequenced using an Illumina HiSeq system in 101-base pair paired-end reads in rapid mode. An average of 23 M paired-end reads were yielded per sample. After trimming sequencing adaptors, low quality reads and bases with a Trimmomatic-0.30(ref. 56) tool, the sequence reads were aligned to a human reference genome (GRCh37/hg19) using Tophat 2.0.11 (bowtie2-2.2.1),57 which adequately aligned the reads to the appropriate location and splice sites in the genome sequence. Files of the gene model annotations and known transcripts were downloaded from the Illumina iGenomes website (http://support.illumina.com/sequencing/sequencing_software/igenome.html), which are necessary for the whole transcriptome alignment with Tophat. The aligned reads were subjected to successive analyses using AvadisNGS 1.6 software (Agilent Technologies). The read counts allocated for each gene and transcript (Ensembl version 2013.04.03) were quantified using a Trimmed Mean of M-value method.58 Statistical analyses between paired groups were performed by paired t-tests. The differentially expressed genes were estimated to be significant if the fold change⩾1.5.
Two hundred and six annotated human pathways were downloaded from GenMAPP (http://www.genmapp.org) and WikiPathways (http://www.wikipathways.org). The collected gene expression data were mapped to these pathways using GenMAPP 2.1. Permutation P values were calculated by MAPPFinder 2.0 to estimate statistically significant pathways having a relative amount of genes with an FC<−2.0 (RC to myxoid).
Identification of miR-135b target mRNAs
To identify the downstream targets of miR-135b, we performed mRNA sequencing and mRNA microarray analyses as described above. We performed in silico analyses to identify potential targets of miRNA using TargetScanHuman 6.0 (http://www.targetscan.org), miRDB (http://www.mirdb.org), PicTar (http://www.pictar.mdc-berlin.de), microRNA.org (http://www.microrna.org) and DIANAmicroT-CDS (http://www.microrna.gr/microT-CDS).
Animal experiments
All animal experiments in this study were performed in compliance with the guidelines of the Institute for Laboratory Animal Research at the National Cancer Center Research Institute (Numbers: T14-003 and A333-14). NOD/Shi-scid, IL-2RγKO Jic (NOG) mice (Central Institute for Experimental Animals, Kawasaki, Kanagawa, Japan) were purchased at 6 weeks of age. Fifty microliters of Matrigel basement membrane matrix (CORNING, Corning, NY, USA) containing 2645-94 GFP-Luc cells (2 × 107) or 2645-94 GFP-Luc LM2 cells (1 × 107) were injected intramuscularlly. To detect the enhancement of tumor size and the appearance of distant metastases, mice were administered 100 mg/kg of D-luciferin (Promega, Madison, WI, USA) intraperitoneally. The signal emerging from tumor cells was visualized and quantified using the IVIS Spectrum imaging system (Caliper Life Science, Hopkinton, MA, USA). Tumor development was monitored once a week. At 3 and 10 weeks after cell injection, tumors and lungs were extracted from the 2645-94 GFP-Luc LM2 and 2645-94 GFP-Luc mice, respectively. The tumor volumes were estimated by the formula V=a2 × b/2, where a and b are the short and long tumor axes, respectively. The cellular density of primary tumor and the number of lung-metastatic foci were evaluated by microscopy. The area of primary site and the focus size were measured by BZ-X analyzer software (Keyence, Osaka, Japan). To establish a high lung-metastatic cell line, the 2645-94 GFP-Luc LM2 cell line, consecutive in vivo lung metastasis selections were performed twice using the 2645-94 GFP-Luc cell line.
Statistics
The data presented in bar graphs are the mean±s.e.m. of at least three independent experiments. The statistical analyses were conducted using Student’s t test (direct comparison), Dunnett's test (multiple comparison) and Fisher’s exact test. We performed receiver-operating characteristic curve analyses using SPSS Statistics Version 21 software (IBM SPSS, Armonk, NY, USA). The optimal cutoff points for the expression levels of miR-135b and THBS2 were determined by the Youden index. The Kaplan–Meier method and the log-rank test were used to compare patient survival. We defined the survival period as the time from diagnosis until recurrence or metastasis, whereas living patients were censored at the time of their last follow-up. P values<0.05 were considered significant.
References
Grosso F, Jones RL, Demetri GD, Judson IR, Blay JY, Le Cesne A et al. Efficacy of trabectedin (ecteinascidin-743) in advanced pretreated myxoid liposarcomas: a retrospective study. Lancet Oncol 2007; 8: 595–602.
Moreau LC, Turcotte R, Ferguson P, Wunder J, Clarkson P, Masri B et al. Myxoid/round cell liposarcoma (MRCLS) revisited: an analysis of 418 primarily managed cases. Ann Surg Oncol 2012; 19: 1081–1088.
Fiore M, Grosso F, Lo Vullo S, Pennacchioli E, Stacchiotti S, Ferrari A et al. Myxoid/round cell and pleomorphic liposarcomas. Cancer 2007; 109: 2522–2531.
Kim H, Lee J, Yi S, Jun H, Choi Y-L, Ahn G et al. Liposarcoma: exploration of clinical prognostic factors for risk based stratification of therapy. BMC Cancer 2009; 9: 205.
Haniball J, Sumathi VP, Kindblom LG, Abudu A, Carter SR, Tillman RM et al. Prognostic factors and metastatic patterns in primary myxoid/round-cell liposarcoma. Sarcoma 2011; 2011: 1–10.
Guan Z, Yu X, Wang H, Wang H, Zhang J, Li G et al. Advances in the targeted therapy of liposarcoma. Onco Targets Ther 2015; 8: 125–136.
Fritchie KJ, Goldblum JR, Tubbs RR, Sun Y, Carver P, Billings SD et al. The expanded histologic spectrum of myxoid liposarcoma with an emphasis on newly described patterns: implications for diagnosis on small biopsy specimens. Am J Clin Pathol 2012; 137: 229–239.
Knight JC, Renwick PJ, Cin PD, Van den Berghe H, Fletcher CDM . Translocation t(12;16)(q13;p11) in myxoid liposarcoma and round cell liposarcoma: molecular and cytogenetic analysis. Cancer Res 1995; 55: 24–27.
Calin GA, Croce CM . MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6: 857–866.
Esquela-Kerscher A, Slack FJ . Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 2006; 6: 259–269.
Borjigin N, Ohno S, Wu W, Tanaka M, Suzuki R, Fujita K et al. TLS-CHOP represses miR-486 expression, inducing upregulation of a metastasis regulator PAI-1 in human myxoid liposarcoma. Biochem Biophys Res Commun 2012; 427: 355–360.
Lee DH, Amanat S, Goff C, Weiss LM, Said JW, Doan NB et al. Overexpression of miR-26a-2 in human liposarcoma is correlated with poor patient survival. Oncogenesis 2013; 2: e47.
Renner M, Czwan E, Hartmann W, Penzel R, Brors B, Eils R et al. MicroRNA profiling of primary high-grade soft tissue sarcomas. Genes Chromosomes Cancer 2012; 51: 982–996.
Gits CM, van Kuijk PF, Jonkers MB, Boersma AW, Smid M, van Ijcken WF et al. MicroRNA expression profiles distinguish liposarcoma subtypes and implicate miR-145 and miR-451 as tumor suppressors. Int J Cancer 2014; 135: 348–361.
Yang Z, Kyriakides TR, Bornstein P . Matricellular proteins as modulators of cell-matrix interactions: adhesive defect in thrombospondin 2-null fibroblasts is a consequence of increased levels of matrix metalloproteinase-2. Mol Biol Cell 2000; 11: 3353–3364.
Yang Z, Strickland DK, Bornstein P . Extracellular matrix metalloproteinase 2 levels are regulated by the low density lipoprotein-related scavenger receptor and thrombospondin 2. J Biol Chem 2001; 276: 8403–8408.
Bornstein P . Thrombospondins as matricellular modulators of cell function. J Clin Invest 2001; 107: 929–934.
Dei Tos AP . Liposarcomas: diagnostic pitfalls and new insights. Histopathology 2014; 64: 38–52.
Pérez-Losada J, Pintado B, Gutiérrez-Adán A, Flores T, Bañares-González B, del Campo JC et al. The chimeric FUS/TLS-CHOP fusion protein specifically induces liposarcomas in transgenic mice. Oncogene 2000; 19: 2413–2422.
Pérez-Losada J, Sánchez-Martín M, Rodríguez-García MA, Pérez-Mancera PA, Pintado B, Flores T et al. Liposarcoma initiated by FUS/TLS-CHOP: the FUS/TLS domain plays a critical role in the pathogenesis of liposarcoma. Oncogene 2000; 19: 6015–6022.
Smith TA, Easley KA, Goldblum JR, Flynn JM, Schwend RM, Aljassir F et al. Myxoid/round cell liposarcoma of the extremities. A clinicopathologic study of 29 cases with particular attention to extent of round cell liposarcoma. Am J Surg Pathol 1996; 20: 171–180.
De Cecco L, Negri T, Brich S, Mauro V, Bozzi F, Dagrada G et al. Identification of a gene expression driven progression pathway in myxoid liposarcoma. Oncotarget 2014; 5: 5965–5977.
Matushansky I, Hernando E, Socci ND, Matos T, Mills J, Edgar MA et al. A developmental model of sarcomagenesis defines a differentiation-based classification for liposarcomas. Am J Pathol 2008; 172: 1069–1080.
Demicco EG, Torres KE, Ghadimi MP, Colombo C, Bolshakov S, Hoffman A et al. Involvement of the PI3K/Akt pathway in myxoid/round cell liposarcoma. Mod Pathol 2011; 25: 212–221.
Nabeshima A, Matsumoto Y, Fukushi J, Iura K, Matsunobu T, Endo M et al. Tumour-associated macrophages correlate with poor prognosis in myxoid liposarcoma and promote cell motility and invasion via the HB-EGF-EGFR-PI3K/Akt pathways. Br J Cancer 2015; 112: 547–555.
Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res 2008; 68: 5795–5802.
Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell 2014; 25: 469–483.
Wu W, Wang Z, Yang P, Yang J, Liang J, Chen Y et al. MicroRNA-135b regulates metastasis suppressor 1 expression and promotes migration and invasion in colorectal cancer. Mol Cell Biochem 2014; 388: 249–259.
He Y, Wang J, Wang J, Yung VY-W, Hsu E, Li A et al. MicroRNA-135b regulates apoptosis and chemoresistance in colorectal cancer by targeting large tumor suppressor kinase 2. Am J Cancer Res 2015; 5: 1382–1395.
Lin CW, Chang YL, Chang YC, Lin JC, Chen CC, Pan SH et al. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat Commun 2013; 4: 1877.
Munding JB, Adai AT, Maghnouj A, Urbanik A, Zöllner H, Liffers ST et al. Global microRNA expression profiling of microdissected tissues identifies miR-135b as a novel biomarker for pancreatic ductal adenocarcinoma. Int J Cancer 2012; 131: E86–E95.
Li Y, Xu D, Bao C, Zhang Y, Chen D, Zhao F . MicroRNA-135b, a HSF1 target, promotes tumor invasion and metastasis by regulating RECK and EVI5 in hepatocellular carcinoma. Oncotarget 2015; 6: 2421–2433.
Pei H, Jin Z, Chen S, Sun X, Yu J, Guo W . MiR-135b promotes proliferation and invasion of osteosarcoma cells via targeting FOXO1. Mol Cell Biochem 2015; 400: 245–252.
Sun R, Wu J, Chen Y, Lu M, Zhang S, Lu D et al. Down regulation of Thrombospondin2 predicts poor prognosis in patients with gastric cancer. Mol Cancer 2014; 13: 225.
Matos AR, Coutinho-Camillo CM, Thuler LC, Fonseca FP, Soares FA, Silva EA et al. Expression analysis of thrombospondin 2 in prostate cancer and benign prostatic hyperplasia. Exp Mol Pathol 2013; 94: 438–444.
Tokunaga T, Nakamura M, Oshika Y, Abe Y, Ozeki Y, Fukushima Y et al. Thrombospondin 2 expression is correlated with inhibition of angiogenesis and metastasis of colon cancer. Br J Cancer 1999; 79: 354–359.
Nakamura M, Oida Y, Abe Y, Yamazaki H, Mukai M, Matsuyama M et al. Thrombospondin-2 inhibits tumor cell invasion through the modulation of MMP-9 and uPA in pancreatic cancer cells. Mol Med Rep 2008; 1: 423–427.
Chijiwa T, Abe Y, Ikoma N, Yamazaki H, Tsukamoto H, Suemizu H et al. Thrombospondin 2 inhibits metastasis of human malignant melanoma through microenvironment-modification in NOD/SCID/gammaCnull (NOG) mice. Int J Oncol 2009; 34: 5–13.
Chen K, Rajewsky N . Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet 2006; 38: 1452–1456.
Saunders MA, Liang H, Li W . Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA 2007; 104: 3300–3305.
Sethupathy P, Collins FS . MicroRNA target site polymorphisms and human disease. Trends Genet 2008; 24: 489–497.
Sato H, Takino T . Coordinate action of membrane-type matrix metalloproteinase-1 (MT1-MMP) and MMP-2 enhances pericellular proteolysis and invasion. Cancer Sci 2010; 101: 843–847.
Oda Y, Yamamoto H, Takahira T, Kobayashi C, Kawaguchi K, Tateishi N et al. Frequent alteration of p16INK4a/p14ARF and p53 pathways in the round cell component of myxoid/round cell liposarcoma: p53 gene alterations and reduced p14ARF expression both correlate with poor prognosis. J Pathol 2005; 207: 410–421.
Schneider-Stock R, Boltze C, Jäger V, Epplen J, Landt O, Peters B et al. Elevated telomerase activity,c-MYC-, and hTERT mRNA expression: association with tumour progression in malignant lipomatous tumours. J Pathol 2003; 199: 517–525.
Chou J, Shahi P, Werb Z . MicroRNA-mediated regulation of the tumor microenvironment. Cell Cycle 2013; 12: 3262–3271.
Smith SM, Coleman J, Bridge JA, Iwenofu OH . Molecular diagnostics in soft tissue sarcomas and gastrointestinal stromal tumors. J Surg Oncol 2015; 111: 520–531.
Fujiwara T, Kunisada T, Takeda K, Uotani K, Yoshida A, Ochiya T et al. MicroRNAs in soft tissue sarcomas: overview of the accumulating evidence and importance as novel biomarkers. Biomed Res Int 2014; 2014: 592868.
Kosaka N, Iguchi H, Ochiya T . Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010; 101: 2087–2092.
Schwarzenbach H, Nishida N, Calin GA, Pantel K . Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 2014; 11: 145–156.
Taylor DD, Gercel-Taylor C . MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008; 110: 13–21.
Zen K, Zhang CY . Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev 2012; 32: 326–348.
Tsukamoto O, Miura K, Mishima H, Abe S, Kaneuchi M, Higashijima A et al. Identification of endometrioid endometrial carcinoma-associated microRNAs in tissue and plasma. Gynecol Oncol 2014; 132: 715–721.
Wu CW, Ng SC, Dong Y, Tian L, Ng SS, Leung WW et al. Identification of microRNA-135b in stool as a potential noninvasive biomarker for colorectal cancer and adenoma. Clin Cancer Res 2014; 20: 2994–3002.
Aman P, Ron D, Mandahl N, Fioretos T, Heim S, Arheden K et al. Rearrangement of the transcription factor gene CHOP in myxoid liposarcomas with t(12;16)(q13;p11). Genes Chromosomes Cancer 1992; 5: 278–285.
Thelin-Järnum S, Lassen C, Panagopoulos I, Mandahl N, Aman P . Identification of genes differentially expressed in TLS-CHOP carrying myxoid liposarcomas. Int J Cancer 1999; 83: 30–33.
Bolger AM, Lohse M, Usadel B . Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–2120.
Langmead B, Salzberg SL . Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9: 357–359.
Robinson MD, Oshlack A . A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010; 11: R25.
Acknowledgements
We thank Masahiko Kuroda for kindly providing MLS cell lines. We also acknowledge Luc Gailhouste for his careful review of the manuscript. This work was supported in part by a grant-in-aid for the third term of a comprehensive 10-year strategy for cancer control, a grant-in-aid for scientific research on priority area cancers from the Ministry of Education, Culture, Sports, Science, and Technology, and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NiBio), the Japan Society for the Promotion of Science through the Funding Program for world-leading innovative R&D on science and technology (FIRST Program) initiated by the Council for Science and Technology Policy, and a grant-in-aid for Scientific Research on Innovative Areas (functional machinery for non-coding RNAs) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Nezu, Y., Hagiwara, K., Yamamoto, Y. et al. miR-135b, a key regulator of malignancy, is linked to poor prognosis in human myxoid liposarcoma. Oncogene 35, 6177–6188 (2016). https://doi.org/10.1038/onc.2016.157
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.157
This article is cited by
-
Role of FUS-CHOP in Myxoid Liposarcoma via miR-486/CDK4 Axis
Biochemical Genetics (2022)
-
Molecular mechanisms underpinning sarcomas and implications for current and future therapy
Signal Transduction and Targeted Therapy (2021)
-
Molecular signatures of tumor progression in myxoid liposarcoma identified by N-glycan mass spectrometry imaging
Laboratory Investigation (2020)
-
Lnc SMAD5-AS1 as ceRNA inhibit proliferation of diffuse large B cell lymphoma via Wnt/β-catenin pathway by sponging miR-135b-5p to elevate expression of APC
Cell Death & Disease (2019)
-
The MiR-135b–BMAL1–YY1 loop disturbs pancreatic clockwork to promote tumourigenesis and chemoresistance
Cell Death & Disease (2018)