Abstract
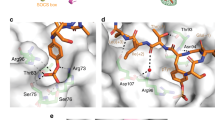
Cbls are RING ubiquitin ligases that attenuate receptor tyrosine kinase (RTK) signal transduction. Cbl ubiquitination activity is stimulated by phosphorylation of a linker helix region (LHR) tyrosine residue. To elucidate the mechanism of activation, we determined the structures of human CBL, a CBL−substrate peptide complex and a phosphorylated-Tyr371-CBL−E2−substrate peptide complex, and we compared them with the known structure of a CBL−E2−substrate peptide complex. Structural and biochemical analyses show that CBL adopts an autoinhibited RING conformation, where the RING's E2-binding surface associates with CBL to reduce E2 affinity. Tyr371 phosphorylation activates CBL by inducing LHR conformational changes that eliminate autoinhibition, flip the RING domain and E2 into proximity of the substrate-binding site and transform the RING domain into an enhanced E2-binding module. This activation is required for RTK ubiquitination. Our results present a mechanism for regulation of c-Cbl's activity by autoinhibition and phosphorylation-induced activation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Pickart, C.M. & Eddins, M.J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 (2004).
Dye, B.T. & Schulman, B.A. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 36, 131–150 (2007).
Duda, D.M. et al. Structural regulation of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 21, 257–264 (2011).
Deshaies, R.J. & Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009).
Li, W. et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle′s dynamics and signaling. PLoS ONE 3, e1487 (2008).
Joazeiro, C.A. et al. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286, 309–312 (1999).
Levkowitz, G. et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 4, 1029–1040 (1999).
Schmidt, M.H. & Dikic, I. The Cbl interactome and its functions. Nat. Rev. Mol. Cell Biol. 6, 907–918 (2005).
Swaminathan, G. & Tsygankov, A.Y. The Cbl family proteins: ring leaders in regulation of cell signaling. J. Cell. Physiol. 209, 21–43 (2006).
Yoon, C.H., Lee, J., Jongeward, G.D. & Sternberg, P.W. Similarity of sli-1, a regulator of vulval development in C. elegans, to the mammalian proto-oncogene c–cbl. Science 269, 1102–1105 (1995).
Peschard, P. et al. Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol. Cell 27, 474–485 (2007).
Kozlov, G. et al. Structural basis for UBA-mediated dimerization of c-Cbl ubiquitin ligase. J. Biol. Chem. 282, 27547–27555 (2007).
Lupher, M.L. Jr., Songyang, Z., Shoelson, S.E., Cantley, L.C. & Band, H. The Cbl phosphotyrosine-binding domain selects a D(N/D)XpY motif and binds to the Tyr292 negative regulatory phosphorylation site of ZAP-70. J. Biol. Chem. 272, 33140–33144 (1997).
Meng, W., Sawasdikosol, S., Burakoff, S.J. & Eck, M.J. Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature 398, 84–90 (1999).
Zheng, N., Wang, P., Jeffrey, P.D. & Pavletich, N.P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533–539 (2000).
Andoniou, C.E., Thien, C.B. & Langdon, W.Y. Tumour induction by activated abl involves tyrosine phosphorylation of the product of the cbl oncogene. EMBO J. 13, 4515–4523 (1994).
Blake, T.J., Shapiro, M., Morse, H.C. III & Langdon, W.Y. The sequences of the human and mouse c–cbl proto-oncogenes show v–cbl was generated by a large truncation encompassing a proline-rich domain and a leucine zipper-like motif. Oncogene 6, 653–657 (1991).
Bisson, S.A., Ujack, E.E. & Robbins, S.M. Isolation and characterization of a novel, transforming allele of the c-Cbl proto-oncogene from a murine macrophage cell line. Oncogene 21, 3677–3687 (2002).
Thien, C.B., Walker, F. & Langdon, W.Y. RING finger mutations that abolish c-Cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol. Cell 7, 355–365 (2001).
Sanada, M. et al. Gain-of-function of mutated C–CBL tumour suppressor in myeloid neoplasms. Nature 460, 904–908 (2009).
Niemeyer, C.M. et al. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat. Genet. 42, 794–800 (2010).
Grand, F.H. et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood 113, 6182–6192 (2009).
Kales, S.C., Ryan, P.E., Nau, M.M. & Lipkowitz, S. Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Res. 70, 4789–4794 (2010).
Kassenbrock, C.K. & Anderson, S.M. Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. J. Biol. Chem. 279, 28017–28027 (2004).
Ryan, P.E., Sivadasan-Nair, N., Nau, M.M., Nicholas, S. & Lipkowitz, S. The N terminus of Cbl-c regulates ubiquitin ligase activity by modulating affinity for the ubiquitin-conjugating enzyme. J. Biol. Chem. 285, 23687–23698 (2010).
Zhang, Y. et al. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics 4, 1240–1250 (2005).
Liu, J., Kimura, A., Baumann, C.A. & Saltiel, A.R. APS facilitates c-Cbl tyrosine phosphorylation and GLUT4 translocation in response to insulin in 3T3–L1 adipocytes. Mol. Cell. Biol. 22, 3599–3609 (2002).
Hu, J. & Hubbard, S.R. Structural characterization of a novel Cbl phosphotyrosine recognition motif in the APS family of adapter proteins. J. Biol. Chem. 280, 18943–18949 (2005).
Ng, C. et al. Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates. EMBO J. 27, 804–816 (2008).
Dominguez, C. et al. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure 12, 633–644 (2004).
Mace, P.D. et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J. Biol. Chem. 283, 31633–31640 (2008).
Brzovic, P.S. et al. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. USA 100, 5646–5651 (2003).
Buchwald, G. et al. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 25, 2465–2474 (2006).
Visser Smit, G.D. et al. Cbl controls EGFR fate by regulating early endosome fusion. Sci. Signal. 2, ra86 (2009).
Ryan, P.E., Davies, G.C., Nau, M.M. & Lipkowitz, S. Regulating the regulator: negative regulation of Cbl ubiquitin ligases. Trends Biochem. Sci. 31, 79–88 (2006).
Saha, A. & Deshaies, R.J. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 (2008).
Siepmann, T.J., Bohnsack, R.N., Tokgoz, Z., Baboshina, O.V. & Haas, A.L. Protein interactions within the N-end rule ubiquitin ligation pathway. J. Biol. Chem. 278, 9448–9457 (2003).
Song, J.J. et al. c-Cbl-mediated degradation of TRAIL receptors is responsible for the development of the early phase of TRAIL resistance. Cell. Signal. 22, 553–563 (2010).
Yokouchi, M. et al. Src-catalyzed phosphorylation of c-Cbl leads to the interdependent ubiquitination of both proteins. J. Biol. Chem. 276, 35185–35193 (2001).
Pufall, M.A. & Graves, B.J. Autoinhibitory domains: modular effectors of cellular regulation. Annu. Rev. Cell Dev. Biol. 18, 421–462 (2002).
Zhang, M. et al. Chaperoned ubiquitylation–crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20, 525–538 (2005).
Du, F., Navarro-Garcia, F., Xia, Z., Tasaki, T. & Varshavsky, A. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc. Natl. Acad. Sci. USA 99, 14110–14115 (2002).
Lopez, J. et al. CARD-mediated autoinhibition of cIAP1′s E3 ligase activity suppresses cell proliferation and migration. Mol. Cell 42, 569–583 (2011).
Chaugule, V.K. et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 30, 2853–2867 (2011).
Yamoah, K. et al. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1′s C-terminal tail. Proc. Natl. Acad. Sci. USA 105, 12230–12235 (2008).
Duda, D.M. et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 (2008).
Gallagher, E., Gao, M., Liu, Y.C. & Karin, M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc. Natl. Acad. Sci. USA 103, 1717–1722 (2006).
Wiesner, S. et al. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130, 651–662 (2007).
Goldenberg, S.J. et al. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119, 517–528 (2004).
Kobashigawa, Y. et al. Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proc. Natl. Acad. Sci. USA 108, 20579–20584 (2011).
Acknowledgements
We would like to thank B. Schulman, F. Kozielski and A. Schuettelkopf for helpful discussions; V. Ulaganathan for computer support; S. Lilla and N. Morrice for mass spectroscopic analyses; W. Clark and A. Keith for in-house DNA sequencing; the Diamond Light Source (DLS) for access to beamlines I02, I03 and I04−1 (mx1229 and mx6683) that contributed to the results presented here; and the ID14−1 beamline at the European Synchrotron Radiation Facility (ESRF) for access and synchrotron support. This work was supported by Cancer Research UK.
Author information
Authors and Affiliations
Contributions
H.D., L.B., G.J.S. and D.T.H. designed, conducted and analyzed in vitro experiments. A.H. designed, conducted and analyzed in vivo experiments. K.H.V. designed and analyzed in vivo experiments. L.B. and D.T.H. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–8 and Supplementary Methods (PDF 7587 kb)
Rights and permissions
About this article
Cite this article
Dou, H., Buetow, L., Hock, A. et al. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat Struct Mol Biol 19, 184–192 (2012). https://doi.org/10.1038/nsmb.2231
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2231
This article is cited by
-
The UAS thioredoxin-like domain of UBXN7 regulates E3 ubiquitin ligase activity of RNF111/Arkadia
BMC Biology (2023)
-
Adipose-specific deletion of the cation channel TRPM7 inhibits TAK1 kinase-dependent inflammation and obesity in male mice
Nature Communications (2023)
-
Structure of UBE2K–Ub/E3/polyUb reveals mechanisms of K48-linked Ub chain extension
Nature Chemical Biology (2022)
-
E3 ligase-inactivation rewires CBL interactome to elicit oncogenesis by hijacking RTK–CBL–CIN85 axis
Oncogene (2021)
-
Depletion of Csk preferentially reduces the protein level of LynA in a Cbl-dependent manner in cancer cells
Scientific Reports (2020)