Key Points
-
The human intestine contains ∼100 trillion bacteria that make essential contributions to human metabolism and establish symbiotic relationships with their hosts. However, these organisms pose an ongoing threat of invasion owing to their enormous numbers.
-
The intestinal immune system has evolved unique immune adaptations that allow it to manage its high bacterial load. These immune mechanisms work together to ensure that commensal bacteria rarely breach the intestinal barrier and that any that do invade are killed rapidly and do not penetrate to systemic sites.
-
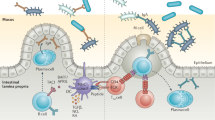
A key element of the mammalian intestinal strategy for maintaining homeostasis with the microbiota is to minimize contact between luminal microorganisms and the intestinal epithelial cell surface. This is accomplished by enhancing the physical barrier through the production of mucus, antimicrobial proteins and IgA.
-
A second layer of intestinal immune protection relies on the rapid detection and killing of bacteria that penetrate the epithelial cell surface. This occurs by several immune mechanisms, including bacterial uptake and phagocytosis by innate immune cells and a complex set of T cell-mediated responses.
-
A third immune barrier is presented by the mesenteric lymph nodes, which constitute an immune 'firewall' limiting penetration by commensal microorganisms to the systemic immune system. This allows induction of adaptive immune responses to resident bacteria to be confined to the mucosal immune compartment.
-
An increasing amount of evidence suggests that inflammatory bowel disease (IBD) arises from dysregulated control of host–microorganism interactions. In support of this hypothesis, several IBD risk alleles compromise intestinal immune mechanisms that maintain homeostasis with the microbiota.
Abstract
Humans harbour nearly 100 trillion intestinal bacteria that are essential for health. Millions of years of co-evolution have moulded this human–microorganism interaction into a symbiotic relationship in which gut bacteria make essential contributions to human nutrient metabolism and in return occupy a nutrient-rich environment. Although intestinal microorganisms carry out essential functions for their hosts, they pose a constant threat of invasion owing to their sheer numbers and the large intestinal surface area. In this Review, we discuss the unique adaptations of the intestinal immune system that maintain homeostatic interactions with a diverse resident microbiota.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
07 April 2015
In figure 4 of the original article, the cytokines that promote the differentiation of T helper 2 (TH2) cells and TH17 cells were included in the wrong order. This has now been corrected in the online HTML and PDF versions of the article. Nature Reviews Immunology apologizes for this error.
References
Mathers, C. D., Boerma, T. & Ma Fat, D. Global and regional causes of death. Br. Med. Bull. 92, 7–32 (2009).
Xu, J. et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076 (2003).
Sonnenburg, J. L. et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959 (2005).
Martens, E. C., Chiang, H. C. & Gordon, J. I. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457 (2008).
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R. & Gordon, J. I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Rev. Microbiol. 6, 776–788 (2008).
Xu, J. & Gordon, J. I. Inaugural article: honor thy symbionts. Proc. Natl Acad. Sci. USA 100, 10452–10459 (2003).
Hooper, L. V. et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291, 881–884 (2001). This paper shows that commensal bacteria manipulate host cell functions and widely influence host biology, revealing the essential nature of the interactions between resident microorganisms and their mammalian hosts.
Hooper, L. V., Stappenbeck, T. S., Hong, C. V. & Gordon, J. I. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nature Immunol. 4, 269–273 (2003).
Stappenbeck, T. S., Hooper, L. V. & Gordon, J. I. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl Acad. Sci. USA 99, 15451–15455 (2002).
He, B. et al. Intestinal bacteria trigger T cell-independent immunoglobulin A2 class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity 26, 812–826 (2007).
Ivanov, I. I. et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 (2008).
Hall, J. A. et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 29, 637–649 (2008).
Stecher, B. et al. Comparison of Salmonella enterica serovar Typhimurium colitis in germfree mice and mice pretreated with streptomycin. Infect. Immun. 73, 3228–3241 (2005).
Klare, I., Werner, G. & Witte, W. Enterococci. Habitats, infections, virulence factors, resistances to antibiotics, transfer of resistance determinants. Contrib. Microbiol. 8, 108–122 (2001).
Benson, A., Pifer, R., Behrendt, C. L., Hooper, L. V. & Yarovinsky, F. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe 6, 187–196 (2009).
Kuwahara, T. et al. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl Acad. Sci. USA 101, 14919–14924 (2004).
Johansson, M. E. et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl Acad. Sci. USA 105, 15064–15069 (2008). This paper clearly visualizes the spatial relationships between the microbiota and the intestinal epithelial cell surface, and it shows that mucus glycoproteins are essential for limiting direct contact between luminal bacteria and epithelial cells.
Van der Sluis, M. et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131, 117–129 (2006).
Celli, J. P. et al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc. Natl Acad. Sci. USA 106, 14321–14326 (2009).
Guerry, P. Campylobacter flagella: not just for motility. Trends Microbiol. 15, 456–461 (2007).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Flo, T. H. et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921 (2004).
Putsep, K. et al. Germ-free and colonized mice generate the same products from enteric prodefensins. J. Biol. Chem. 275, 40478–40482 (2000).
Brandl, K., Plitas, G., Schnabl, B., Dematteo, R. P. & Pamer, E. G. MyD88-mediated signals induce the bactericidal lectin RegIIIγ and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med. 204, 1891–1900 (2007).
Vaishnava, S., Behrendt, C. L., Ismail, A. S., Eckmann, L. & Hooper, L. V. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl Acad. Sci. USA 105, 20858–20863 (2008).
Kobayashi, K. S. et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307, 731–734 (2005).
Meyer-Hoffert, U. et al. Secreted enteric antimicrobial activity localizes to the mucus surface layer. Gut 57, 764–771 (2008).
Boneca, I. G. et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl Acad. Sci. USA 104, 997–1002 (2007).
Guo, L. et al. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95, 189–198 (1998).
Raffatellu, M. et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5, 476–486 (2009).
Suzuki, K. et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl Acad. Sci. USA 101, 1981–1986 (2004).
Macpherson, A. J. et al. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288, 2222–2226 (2000).
Macpherson, A. J. & Uhr, T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303, 1662–1665 (2004). This study shows that DCs harbouring live commensal bacteria are restricted to the mucosal immune compartment by mesenteric lymph nodes, which thus function as an immune firewall that limits systemic penetration of commensal bacteria.
Rescigno, M. et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nature Immunol. 2, 361–367 (2001).
Niess, J. H. et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307, 254–258 (2005).
Fagarasan, S. & Honjo, T. Intestinal IgA synthesis: regulation of front-line body defences. Nature Rev. Immunol. 3, 63–72 (2003).
Lee, S. H., Starkey, P. M. & Gordon, S. Quantitative analysis of total macrophage content in adult mouse tissues. Immunochemical studies with monoclonal antibody F4/80. J. Exp. Med. 161, 475–489 (1985).
Kelsall, B. Recent progress in understanding the phenotype and function of intestinal dendritic cells and macrophages. Mucosal Immunol. 1, 460–469 (2008).
Smythies, L. E. et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J. Clin. Invest. 115, 66–75 (2005).
Sansonetti, P. J. War and peace at mucosal surfaces. Nature Rev. Immunol. 4, 953–964 (2004).
Macpherson, A. J., Geuking, M. B. & McCoy, K. D. Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology 115, 153–162 (2005).
Pull, S. L., Doherty, J. M., Mills, J. C., Gordon, J. I. & Stappenbeck, T. S. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc. Natl Acad. Sci. USA 102, 99–104 (2005).
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004).
Shroff, K. E., Meslin, K. & Cebra, J. J. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immun. 63, 3904–3913 (1995).
Benveniste, J., Lespinats, G. & Salomon, J. Serum and secretory IgA in axenic and holoxenic mice. J. Immunol. 107, 1656–1662 (1971).
Guy-Grand, D. et al. Two gut intraepithelial CD8+ lymphocyte populations with different T cell receptors: a role for the gut epithelium in T cell differentiation. J. Exp. Med. 173, 471–481 (1991).
Macpherson, A. J. & Harris, N. L. Interactions between commensal intestinal bacteria and the immune system. Nature Rev. Immunol. 4, 478–485 (2004).
Barnes, M. J. & Powrie, F. Regulatory T cells reinforce intestinal homeostasis. Immunity 31, 401–411 (2009).
Kuhn, R., Lohler, J., Rennick, D., Rajewsky, K. & Muller, W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274 (1993).
Shull, M. M. et al. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature 359, 693–699 (1992).
Nedjic, J., Aichinger, M., Emmerich, J., Mizushima, N. & Klein, L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature 455, 396–400 (2008).
Powrie, F. et al. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity 1, 553–562 (1994).
Asseman, C., Mauze, S., Leach, M. W., Coffman, R. L. & Powrie, F. An essential role for interleukin 10 in the function of regulatory T cells that inhibit intestinal inflammation. J. Exp. Med. 190, 995–1004 (1999).
Asseman, C., Read, S. & Powrie, F. Colitogenic Th1 cells are present in the antigen-experienced T cell pool in normal mice: control by CD4+ regulatory T cells and IL-10. J. Immunol. 171, 971–978 (2003).
Li, M. O., Wan, Y. Y. & Flavell, R. A. T cell-produced transforming growth factor-β1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity 26, 579–591 (2007).
Cong, Y., Weaver, C. T., Lazenby, A. & Elson, C. O. Bacterial-reactive T regulatory cells inhibit pathogenic immune responses to the enteric flora. J. Immunol. 169, 6112–6119 (2002).
Mazmanian, S. K., Round, J. L. & Kasper, D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008).
Gaboriau-Routhiau, V. et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009).
Neurath, M. F. et al. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn's disease. J. Exp. Med. 195, 1129–1143 (2002).
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O. & Kasper, D. L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005).
Mazmanian, S. K. & Kasper, D. L. The love–hate relationship between bacterial polysaccharides and the host immune system. Nature Rev. Immunol. 6, 849–858 (2006).
Becker, C. et al. Constitutive p40 promoter activation and IL-23 production in the terminal ileum mediated by dendritic cells. J. Clin. Invest. 112, 693–706 (2003).
Gautreaux, M. D., Gelder, F. B., Deitch, E. A. & Berg, R. D. Adoptive transfer of T lymphocytes to T-cell-depleted mice inhibits Escherichia coli translocation from the gastrointestinal tract. Infect. Immun. 63, 3827–3834 (1995).
Weaver, C. T., Harrington, L. E., Mangan, P. R., Gavrieli, M. & Murphy, K. M. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity 24, 677–688 (2006).
Lee, Y. K., Mukasa, R., Hatton, R. D. & Weaver, C. T. Developmental plasticity of Th17 and Treg cells. Curr. Opin. Immunol. 21, 274–280 (2009).
Lee, Y. K. et al. Late developmental plasticity in the T helper 17 lineage. Immunity 30, 92–107 (2009).
Maloy, K. J. et al. CD4+CD25+ TR cells suppress innate immune pathology through cytokine-dependent mechanisms. J. Exp. Med. 197, 111–119 (2003).
Kullberg, M. C. et al. Bacteria-triggered CD4+ T regulatory cells suppress Helicobacter hepaticus-induced colitis. J. Exp. Med. 196, 505–515 (2002).
Kullberg, M. C. et al. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J. Exp. Med. 203, 2485–2494 (2006).
Hue, S. et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J. Exp. Med. 203, 2473–2483 (2006).
Boismenu, R. & Havran, W. L. Modulation of epithelial cell growth by intraepithelial γδ T cells. Science 266, 1253–1255 (1994).
Chen, Y., Chou, K., Fuchs, E., Havran, W. L. & Boismenu, R. Protection of the intestinal mucosa by intraepithelial γδ T cells. Proc. Natl Acad. Sci. USA 99, 14338–14343 (2002).
Groh, V., Steinle, A., Bauer, S. & Spies, T. Recognition of stress-induced MHC molecules by intestinal epithelial γδ T cells. Science 279, 1737–1740 (1998).
Suemizu, H. et al. A basolateral sorting motif in the MICA cytoplasmic tail. Proc. Natl Acad. Sci. USA 99, 2971–2976 (2002).
Ismail, A. S., Behrendt, C. L. & Hooper, L. V. Reciprocal interactions between commensal bacteria and γδ intraepithelial lymphocytes during mucosal injury. J. Immunol. 182, 3047–3054 (2009).
Poussier, P., Ning, T., Banerjee, D. & Julius, M. A unique subset of self-specific intraintestinal T cells maintains gut integrity. J. Exp. Med. 195, 1491–1497 (2002).
Sanos, S. L. et al. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nature Immunol. 10, 83–91 (2009).
Zheng, Y. et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nature Med. 14, 282–289 (2008).
Shiloh, M. U. et al. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity 10, 29–38 (1999). This study shows the essential role of microbicidal mechanisms in containing the commensal intestinal microbiota.
Sansonetti, P. Phagocytosis of bacterial pathogens: implications in the host response. Semin. Immunol. 13, 381–390 (2001).
Gowans, J. L. & Knight, E. J. The route of re-circulation of lymphocytes in the rat. Proc. R. Soc. Lond. B Biol. Sci. 159, 257–282 (1964).
Husband, A. J. & Gowans, J. L. The origin and antigen-dependent distribution of IgA-containing cells in the intestine. J. Exp. Med. 148, 1146–1160 (1978). This is a landmark study of the immune geography of the mucosal immune system, showing the dissemination of IgA+ plasma cells that have been induced in the intestinal mucosa through the lymph and blood.
Pierce, N. F. & Gowans, J. L. Cellular kinetics of the intestinal immune response to cholera toxoid in rats. J. Exp. Med. 142, 1550–1563 (1975).
Nagl, M. et al. Phagocytosis and killing of bacteria by professional phagocytes and dendritic cells. Clin. Diagn. Lab. Immunol. 9, 1165–1168 (2002).
Konrad, A., Cong, Y., Duck, W., Borlaza, R. & Elson, C. O. Tight mucosal compartmentation of the murine immune response to antigens of the enteric microbiota. Gastroenterology 130, 2050–2059 (2006).
Slack, E. et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 325, 617–620 (2009).
Swidsinski, A., Weber, J., Loening-Baucke, V., Hale, L. P. & Lochs, H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43, 3380–3389 (2005).
Hugot, J. P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 (2001).
Ogura, Y. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411, 603–606 (2001). References 88 and 89 show that NOD2 polymorphisms account for a proportion of the genetic risk of Crohn's disease. This was the first of a large number of linked genetic loci to be identified and the suspected dysregulation of host–microorganism mutualism as a cause of IBD was reinforced by the finding that NOD2 senses a bacterial cell wall component.
Wehkamp, J. et al. Reduced Paneth cell α-defensins in ileal Crohn's disease. Proc. Natl Acad. Sci. USA 102, 18129–18134 (2005).
Cadwell, K. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008).
Kaser, A. et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743–756 (2008).
Garabedian, E. M., Roberts, L. J., McNevin, M. S. & Gordon, J. I. Examining the role of Paneth cells in the small intestine by lineage ablation in transgenic mice. J. Biol. Chem. 272, 23729–23740 (1997).
Chapel, H. et al. Common variable immunodeficiency disorders: division into distinct clinical phenotypes. Blood 112, 277–286 (2008).
Notarangelo, L. et al. Primary immunodeficiency diseases: an update from the International Union of Immunological Societies Primary Immunodeficiency Diseases Classification Committee Meeting in Budapest, 2005. J. Allergy Clin. Immunol. 117, 883–896 (2006).
Casanova, J. L. & Abel, L. Primary immunodeficiencies: a field in its infancy. Science 317, 617–619 (2007).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005). This landmark study uses molecular profiling to reveal the diversity of the human microflora and establish the dominant bacterial phylotypes that inhabit the human gastrointestinal tract.
Ogura, Y. et al. Expression of NOD2 in Paneth cells: a possible link to Crohn's ileitis. Gut 52, 1591–1597 (2003).
Inohara, N. et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J. Biol. Chem. 278, 5509–5512 (2003).
Acknowledgements
L.V.H. thanks the students and colleagues from her laboratory for the many discussions that contributed to the ideas in this manuscript. Work in L.V.H.'s laboratory is supported by the Howard Hughes Medical Institute, the US National Institutes of Health (DK070855), the Burroughs Wellcome Foundation and the Crohn's and Colitis Foundation. A.M. acknowledges K. McCoy, E. Slack, S. Hapfelmeier, M. Stoehl and M. Geuking.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Microbiota
-
The microorganisms that are harboured by normal, healthy individuals. These microorganisms live in the digestive tract and at other body sites.
- Sepsis
-
A systemic response to severe infection or tissue damage, leading to a hyperactive and unbalanced network of pro-inflammatory mediators. Vascular permeability, cardiac function and metabolic balance are affected, resulting in tissue necrosis, multi-organ failure and death.
- Metagenome
-
All the genetic material present in a population of microorganisms, consisting of the genomes of many individual organisms.
- Goblet cell
-
A mucus-producing cell found in the epithelial cell lining of the intestine and lungs.
- Defensin
-
A class of antimicrobial peptide that has activity against Gram-positive and Gram-negative bacteria, fungi and viruses. α-defensins are produced by intestinal Paneth cells and neutrophils, and β-defensins are expressed by most epithelial cells.
- C-type lectin
-
An animal receptor protein that binds to carbohydrates, frequently in a Ca2+-dependent manner. The binding activity of C-type lectins is based on the structure of the carbohydrate-recognition domain, which is highly conserved among members of this family.
- Paneth cells
-
A specialized epithelial cell lineage that produces most of the antimicrobial proteins in the small intestine.
- Peyer's patches
-
Groups of lymphoid nodules present in the small intestine (usually the ileum). They occur massed together on the intestinal wall, opposite the line of attachment of the mesentery. Peyer's patches consist of a dome area, B cell follicles and interfollicular T cell areas. High endothelial venules are present mainly in the interfollicular areas.
- Lamina propria
-
Connective tissue that underlies the epithelium of the mucosa and contains various myeloid and lymphoid cells, including macrophages, dendritic cells, T cells and B cells.
- Plasma cell
-
A non-dividing, terminally differentiated, immobile antibody-secreting cell of the B cell lineage.
- Transcytosis
-
Process of transport of material across a cell monolayer by uptake on one side of the cell into a coated vesicle, which might then be sorted through the trans-Golgi network and transported to the opposite side of the cell.
- Germ-free mouse
-
A mouse that is born and raised in isolators, without exposure to microorganisms.
- Germinal centre
-
Located in peripheral lymphoid tissues (for example, the spleen), these structures are sites of B cell proliferation and selection for clones that produce antigen-specific antibodies of higher affinity.
- Recombination-activating gene
-
(Rag). A gene expressed by developing lymphocytes. Mice that are deficient for either Rag1 or Rag2 fail to produce B or T cells owing to a developmental block in the gene rearrangement that is necessary for antigen receptor expression.
- Severe combined immunodeficiency
-
(SCID). A phenotype of mice with a defect in DNA recombination. SCID mice lack B and T cells and do not reject tissue grafts from allogeneic and xenogeneic sources.
- Intraepithelial CD8αα+ T cell
-
A type of T cell that is found in the intestinal epithelium. The CD8 molecule that they express is a homodimer of CD8α, rather than the CD8αβ heterodimer that is expressed by conventional CD8+ T cells in the lymph nodes. It has been proposed that these cells are self-reactive T cells that have regulatory properties.
- Lymphoid-tissue inducer cell
-
(LTi cell). A cell that is present in developing lymph nodes, Peyer's patches and nasopharynx-associated lymphoid tissue. LTi cells are required for the development of these lymphoid organs and are characterized by expression of the transcription factor retinoic acid receptor-related orphan receptor-γt (RORγt), interleukin-7 receptor-α and lymphotoxin-α1β;2.
- Specific pathogen-free (SPF) mice
-
Mice kept in specific vivarium conditions whereby a number of pathogens are excluded or eradicated from the colony. These animals are maintained in the absence of most of the known chronic and latent persistent pathogens. Although this enables better control of experimental conditions related to immunity and infection, it also sets apart such animal models from pathogen-exposed humans or non-human primates, whose immune systems are in constant contact with potential pathogens.
Rights and permissions
About this article
Cite this article
Hooper, L., Macpherson, A. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 10, 159–169 (2010). https://doi.org/10.1038/nri2710
Issue Date:
DOI: https://doi.org/10.1038/nri2710
This article is cited by
-
Current perspectives on fecal microbiota transplantation in inflammatory bowel disease
Indian Journal of Gastroenterology (2024)
-
The long-term gut bacterial signature of a wild primate is associated with a timing effect of pre- and postnatal maternal glucocorticoid levels
Microbiome (2023)
-
Colonization resistance is dispensable for segregation of oral and gut microbiota
BMC Medical Genomics (2023)
-
Daily fluctuation of colonic microbiome in response to nutrient substrates in a pig model
npj Biofilms and Microbiomes (2023)
-
White spot syndrome virus impact on the expression of immune genes and gut microbiome of black tiger shrimp Penaeus monodon
Scientific Reports (2023)