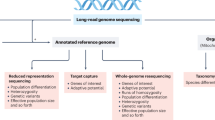
Key Points
-
We will soon have complete genome sequences from thousands of species. This coming explosion of information will transform our understanding of the amount, distribution and functional significance of genetic variation in natural populations.
-
We identify those problems in conservation biology in which genomics will be most valuable in providing new insights and understanding. We also provide guidelines as to which new genomics approaches will be most appropriate for the different problems in conservation that can benefit from genetic analysis.
-
The most straightforward contribution of genomics to conservation will be to enormously increase the precision and accuracy of estimation of crucial parameters that require neutral loci (for example, effective population size and migration rate).
-
Genomic approaches can address important questions about the molecular basis and genetic architecture of inbreeding depression. Recent work indicates that the intensity of inbreeding depression can differ greatly depending on which specific individuals are founders. This suggests that the genetic load is unevenly spread among founder genomes and supports the notion that inbreeding depression sometimes results from major effects at a few loci.
-
Anthropogenic challenges affect a wide range of species and habitats. Genomic approaches will allow the identification of adaptive genetic variation related to key traits for the response to climate change, such as phenology or drought tolerance, so that management may focus on maintaining adaptive genetic potential. The use of genomics to monitor genetic change caused by the harvesting of animals by humans could be extremely important because early detection of potentially harmful genetic change will maximize our ability to implement management to limit or reverse the effects before substantial or irreversible changes occur.
-
Genomics provides exciting opportunities to assess differential rates of introgression across different genomic regions following hybridization between native and introduced species. The differential introgression rates of genomic regions raise some difficult issues with regards to treating hybridized populations in conservation and bring into question the efficacy of using a few (that is, ten or so) neutral markers to detect hybridization.
-
Genomic tools will assist the management of ex situ populations and reintroductions by providing increased precision and accuracy of estimates of neutral population genetic parameters and by identifying specific loci of importance, which are essential for selecting select founder individuals.
-
There is increasing evidence that epigenetic processes can be important following hybridization. Therefore, an epigenetics perspective might be important for understanding the effects of hybridization and predicting outbreeding depression.
-
Improved basic scientific understanding through genomics will not necessarily lead to improved conservation. For example, understanding the relationship between genetic variation and fitness itself will not be sufficient to improve our estimates of population viability. Understanding the connections between individual fitness and population growth rates is perhaps the most important and difficult future challenge facing conservation genetics.
Abstract
We will soon have complete genome sequences from thousands of species, as well as from many individuals within species. This coming explosion of information will transform our understanding of the amount, distribution and functional significance of genetic variation in natural populations. Now is a crucial time to explore the potential implications of this information revolution for conservation genetics and to recognize limitations in applying genomic tools to conservation issues. We identify and discuss those problems for which genomics will be most valuable for curbing the accelerating worldwide loss of biodiversity. We also provide guidance on which genomics tools and approaches will be most appropriate to use for different aspects of conservation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ellegren, H. & Sheldon, B. C. Genetic basis of fitness differences in natural populations. Nature 452, 169–175 (2008). An important paper that reviews current understanding of the molecular basis of fitness differences between individuals in natural populations.
Slate, J. et al. Gene mapping in the wild with SNPs: guidelines and future directions. Genetica 136, 97–107 (2009).
Kohn, M. H., Murphy, W. J., Ostrander, E. A. & Wayne, R. K. Genomics and conservation genetics. Trends Ecol. Evol. 21, 629–637 (2006).
Pertoldi, C. et al. Genome variability in European and American bison detected using the BovineSNP50 BeadChip. Conserv. Genet. 11, 627–634 (2010).
Thomson, R. C., Wang, I. J. & Johnson, J. R. Genome-enabled development of DNA markers for ecology, evolution and conservation. Mol. Ecol. 19, 2184–2195 (2010).
Kerstens, H. et al. Large scale single nucleotide polymorphism discovery in unsequenced genomes using second generation high throughput sequencing technology: applied to turkey. BMC Genomics 10, 479 (2009).
van Bers, N. E. M. et al. Genome-wide SNP detection in the great tit Parus major using high throughput sequencing. Mol. Ecol. 19, 89–99 (2010).
DeLong, E. F. The microbial ocean from genomes to biomes. Nature 459, 200–206 (2009). A review of metagenomics in marine systems, including transcriptomic and functional approaches linking microbial genomes to ecosystem processes.
Dinsdale, E. A. et al. Functional metagenomic profiling of nine biomes. Nature 452, 629–634 (2008).
Vega Thurber, R. L. et al. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc. Natl Acad. Sci. USA 47, 18413–18418 (2008).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Nielsen, E. E., Hemmer-Hansen, J., Larsen, P. F. & Bekkevold, D. Population genomics of marine fishes: identifying adaptive variation in space and time. Mol. Ecol. 18, 3128–3150 (2009).
Murchison, E. P. et al. The Tasmanian devil transcriptome reveals Schwann cell origins of a clonally transmissible cancer. Science 327, 84–87 (2010).
Avise, J. Perspective: conservation genetics enters the genomics era. Conserv. Genet. 11, 665–669 (2010).
Ouborg, N. J., Pertoldi, C., Loeschcke, V., Bijlsma, R. & Hedrick, P. W. Conservation genetics in transition to conservation genomics. Trends Genet. 26, 177–187 (2010).
Primmer, C. R. From conservation genetics to conservation genomics. Ann. N. Y. Acad. Sci. 1162, 357–368 (2009).
Romanov, M. N. et al. The value of avian genomics to the conservation of wildlife. BMC Genomics 10, S10 (2010).
Allendorf, F. W. & Seeb, L. W. Concordance of genetic divergence among sockeye salmon populations at allozyme, nuclear DNA, and mitochondrial DNA markers. Evolution 54, 640–651 (2000).
Luikart, G. H., England, P., Tallmon, D. A., Jordan, S. & Taberlet, P. The power and promise of population genomics: from genotyping to genome-typing. Nature Rev. Genet. 4, 981–994 (2003).
Storz, J. F. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol. Ecol. 14, 671–688 (2005).
Giger, T. et al. Life history shapes gene expression in salmonids. Curr. Biol. 16, R281–R282 (2006).
Landry, P.-A., Koskinen, M. T. & Primmer, C. R. Deriving evolutionary relationships among populations using microsatellites and (δμ)2: all loci are equal, but some are more equal than others. Genetics 161, 1339–1347 (2002).
Nordborg, M. Structured coalescent processes on different time scales. Genetics 146, 1501–1514 (1997).
Beaumont, M. A. Detecting population expansion and decline using microsatellites. Genetics 153, 2013–2029 (1999).
Beerli, P. & Felsenstein, J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Natl Acad. Sci. USA 98, 4563–4568 (2001).
Skare, O., Sheehan, N. & Egeland, T. Identification of distant family relationships. Bioinformatics 25, 2376–2382 (2009).
Browning, S. R. & Weir, B. S. Population structure with localized haplotype clusters. Genetics 10 May 2010 (doi:10.1534/genetics.110.116681).
Lecis, R. et al. Bayesian analyses of admixture in wild and domestic cats (Felis silvestris) using linked microsatellite loci. Mol. Ecol. 15, 119–131 (2006).
Anderson, C. & Meikle, D. Genetic estimates of immigration and emigration rates in relation to population density and forest patch area in Peromyscus leucopus. Conserv. Genet. 5 Feb 2010 (doi:10.1007/s10592-009-0033-8).
Bollback, J. P., York, T. L. & Nielsen, R. Estimation of 2Nes from temporal allele frequency data. Genetics 179, 497–502 (2008).
Wang, J. & Santure, A. W. Parentage and sibship inference from multilocus genotype data under polygamy. Genetics 181, 1579–1594 (2009).
Glaubitz, J. C., Rhodes, O. E. & DeWoody, J. A. Prospects for inferring pairwise relationships with single nucleotide polymorphisms. Mol. Ecol. 12, 1039–1047 (2003).
Jones, O. R. & Wang, J. Molecular marker-based pedigrees for animal conservation biologists. Anim. Conserv. 13, 26–34 (2010).
Keinan, A. & Reich, D. Human population differentiation is strongly correlated with local recombination rate. PLoS Genet. 6, e1000886 (2010).
Santure, A. W. et al. On the use of large marker panels to estimate inbreeding and relatedness: empirical and simulation studies of a pedigreed zebra finch population typed at 771 SNPs. Mol. Ecol. 19, 1439–1451 (2010).
Smouse, P. How many SNPs are enough? Mol. Ecol. 19, 1265–1266 (2010).
Pemberton, J. M. Wild pedigrees: the way forward. Proc. Biol. Sci. 275, 613–621 (2008). A persuasive argument for the value of constructing pedigrees in wild populations to investigate major issues in evolutionary biology, including the genetic architecture of traits, inbreeding depression and inbreeding avoidance.
Sham, P., Cherny, S. & Purcell, S. Application of genome-wide SNP data for uncovering pairwise relationships and quantitative trait loci. Genetica 136, 237–243 (2009).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Hubisz, M. J., Falush, D., Stephens, M. & Pritchard, J. K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 9, 1322–1332 (2009).
Rosenberg, N. A. et al. Genetic structure of human populations. Science 298, 2381–2385 (2002).
Wilson, G. A. & Rannala, B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163, 1177–1191 (2003).
Faubet, P., Waples, R. S. & Gaggiotti, O. E. Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol. Ecol. 16, 1149–1166 (2007).
Rannala, B. & Mountain, J. L. Detecting immigration by using multilocus genotypes. Proc. Natl Acad. Sci. USA 94, 9197–9201 (1997).
Holderegger, R. & Wagner, H. H. Landscape genetics. BioScience 58, 199–207 (2008). An integrated review of the emerging field of landscape genetics.
Hagenblad, J., Olsson, M., Parker, H. G., Ostrander, E. A. & Ellegren, H. Population genomics of the inbred Scandinavian wolf. Mol. Ecol. 18, 1341–1351 (2009).
Casellas, J., Varona, L., Ibanez-Scriche, N., Quintanilla, R. & Noguera, J. L. Skew distribution of founder-specific inbreeding depression effects on the longevity of landrace sows. Genet. Res. 90, 499–508 (2008).
Lacy, R. C., Alaks, G. & Walsh, A. Hierarchical analysis of inbreeding depression in Peromyscus polionotus. Evolution 50, 2187–2200 (1996).
Casellas, J., Piedrafita, J., Caja, G. & Varona, L. Analysis of founder-specific inbreeding depression on birth weight in Ripollesa lambs. J. Anim. Sci. 87, 72–79 (2009).
Charlier, C. et al. Highly effective SNP-based association mapping and management of recessive defects in livestock. Nature Genet. 40, 449–454 (2008).
Vermeulen, C. J., Bijlsma, R. & Loeschcke, V. QTL mapping of inbreeding-related cold sensitivity and conditional lethality in Drosophila melanogaster. J. Evol. Biol. 21, 1236–1244 (2008).
Kristensen, T. N., Pedersen, K. S., Vermeulen, C. J. & Loeschcke, V. Research on inbreeding in the 'omic' era. Trends Ecol. Evol. 25, 44–52 (2010). An important evaluation of the use of new technologies to understand the genetic basis of inbreeding depression.
Roach, J. C. et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328, 636–639 (2010).
Nielsen, R. Molecular signatures of natural selection. Annu. Rev. Genet. 39, 197–218 (2005).
Oleksyk, T. K., Smith, M. W. & O'Brien, S. J. Genome-wide scans for footprints of natural selection. Philos. Trans. R. Soc. Lond. B 365, 185–205 (2010).
Garrigan, D. & Hedrick, P. W. Detecting adaptive molecular polymorphism: lessons from the MHC. Evolution 57, 1707–1722 (2003).
Antao, T., Lopes, A., Lopes, R., Beja-Pereira, A. & Luikart, G. LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics 9, 323 (2008).
Beaumont, M. A. & Balding, D. J. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 13, 969–980 (2004).
Beaumont, M. A. & Nichols, R. A. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. B 263, 1619–1626 (1996).
Foll, M. & Gaggiotti, O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180, 977–993 (2008).
Makinen, H. S., Cano, J. M. & Merilä, J. Identifying footprints of directional and balancing selection in marine and freshwater three-spined stickleback (Gasterosteus aculeatus) populations. Mol. Ecol. 17, 3565–3582 (2008).
Namroud, M.-C., Beaulieu, J., Juge, N., Laroche, J. & Bousquet, J. Scanning the genome for gene single nucleotide polymorphisms involved in adaptive population differentiation in white spruce. Mol. Ecol. 17, 3599–3613 (2008).
Nielsen, E. E. et al. Genomic signatures of local directional selection in a high gene flow marine organism; the Atlantic cod (Gadus morhua). BMC Evol. Biol. 9, 276 (2009).
Oetjen, K. & Reusch, T. B. H. Genome scans detect consistent divergent selection among subtidal vs. intertidal populations of the marine angiosperm Zostera marina. Mol. Ecol. 16, 5156–5167 (2007).
Vasemägi, A., Nilsson, J. & Primmer, C. R. Expressed sequence tag-linked microsatellites as a source of gene-associated polymorphisms for detecting signatures of divergence selection in Atlantic salmon (Salmo salar L.). Mol. Biol. Evol. 22, 1067–1076 (2005).
Hohenlohe, P. A. et al. Population genomic analysis of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 6, e1000862 (2010). One of the first papers to use genomic scans of thousands of markers to understand the genetic basis of adaptation in natural populations.
Wilding, C. S., Butlin, R. K. & Grahame, J. Differential gene exchange between parapatric morphs of Littorina saxatilis detected using AFLP markers. J. Evol. Biol. 14, 611–619 (2001).
Pennings, P. S. & Hermisson, J. Soft sweeps II — molecular population genetics of adaptation from recurrent mutation or migration. Mol. Biol. Evol. 23, 1076–1084 (2006).
Voight, B. F., Kudaravalli, S., Wen, X. & Pritchard, J. K. A map of recent positive selection in the human genome. PLoS Biol. 4, e72 (2006).
Allendorf, F. W. & Hard, J. J. Human-induced evolution caused by unnatural selection through harvest of wild animals. Proc. Natl Acad. Sci. USA 106, 9987–9994 (2009).
Allendorf, F. W., England, P. R., Luikart, G., Ritchie, P. A. & Ryman, N. Genetic effects of harvest on wild animal populations. Trends Ecol. Evol. 23, 327–337 (2008).
Frankham, R. Where are we in conservation genetics and where do we need to go? Conserv. Genet. 11, 661–663 (2010).
Allendorf, F. W., Leary, R. F., Spruell, P. & Wenburg, J. K. The problems with hybrids: setting conservation guidelines. Trends Ecol. Evol. 16, 613–622 (2001).
Levin, D. A., Franciscoortega, J. & Jansen, R. K. Hybridization and the extinction of rare plant species. Conserv. Biol. 10, 10–16 (1996).
Crandall, K. A., Binindaemonds, O. R. P., Mace, G. M. & Wayne, R. K. Considering evolutionary processes in conservation biology. Trends Ecol. Evol. 15, 290–295 (2000).
Hedrick, P. W., Parker, K. M. & Lee, R. N. Using microsatellite and MHC variation to identify species, ESUs, and MUs in the endangered Sonoran topminnow. Mol. Ecol. 10, 1399–1412 (2001).
Bonin, A., Nicole, F., Pompanon, F., Miaud, C. & Taberlet, P. Population adaptive index: a new method to help measure intraspecific genetic diversity and prioritize populations for conservation. Conserv. Biol. 21, 697–708 (2007).
Coop, G. et al. The role of geography in human adaptation. PLoS Genet. 5, e1000500 (2009).
Frazer, K. A., Murray, S. S., Schork, N. J. & Topol, E. J. Human genetic variation and its contribution to complex traits. Nature Rev. Genet. 10, 241–252 (2009).
Hansen, M. M. Expression of interest: transcriptomics and the designation of conservation units. Mol. Ecol. 19, 1757–1759 (2010).
Tymchuk, W. V., O'Reilly, P., Bittman, J., MacDonald, D. & Schulte, P. Conservation genomics of Atlantic salmon: variation in gene expression between and within regions of the Bay of Fundy. Mol. Ecol. 19, 1842–1859 (2010).
Cornuet, J.-M., Piry, S., Luikart, G., Estoup, A. & Solignac, M. New methods employing multilocus genotypes for selecting or excluding populations as origins of individuals. Genetics 153, 1989–2000 (1999).
Witherspoon, D. J. et al. Genetic similarities within and between human populations. Genetics 176, 351–359 (2007).
Halbert, N. D. & Derr, J. N. A comprehensive evaluation of cattle introgression into US federal bison herds. J. Hered. 98, 1–12 (2007).
Allendorf, F. W. et al. Cutthroat trouth hybridization and the U. S. Endangered Species Act: one species, two policies. Conserv. Biol. 19, 1326–1328 (2005).
Campton, D. E. & Kaeding, L. R. Westslope cutthroat trout, hybridization, and the U. S. Endangered Species Act. Conserv. Biol. 19, 1323–1325 (2005).
Marris, E. The genome of the American west. Nature 457, 950–952 (2009).
Payseur, B. A. Using differential introgression in hybrid zones to identify genomic regions involved in speciation. Mol. Ecol. Resour. 10, 806–820 (2010).
Fitzpatrick, B. M. et al. Rapid spread of invasive genes into a threatened native species. Proc. Natl Acad. Sci. USA 107, 3606–3610 (2010). A fascinating study showing that natural selection can rapidly accelerate the rate of introgression for certain regions of the genome from introduced into native species.
Edmands, S. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 16, 463–475 (2007).
Shendure, J. & Ji, H. Next-generation DNA sequencing. Nature Biotech. 26, 1135–1145 (2008).
Hoffmann, A. A. & Rieseberg, L. H. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu. Rev. Ecol. Evol. Syst. 39, 21–42 (2008). An important synthesis that evaluates the importance of chromosomal inversions in population genetics and evolution using modern molecular approaches.
Muhlfeld, C. C. et al. Hybridization rapidly reduces fitness of a native trout in the wild. Biol. Lett. 5, 328–331 (2009).
Ljungqvist, M., Åkesson, M. & Hansson, B. Do microsatellites reflect genome-wide genetic diversity in natural populations? A comment on Väli. et al. (2008). Mol. Ecol. 19, 851–855 (2010).
Miller, W., Wright, S. J., Zhang, Y., Schuster, S. C. & Hayes, V. M. Optimization methods for selecting founder individuals for captive breeding or reintroduction of endangered species. Pac. Symp. Biocomput. 2010, 43–53 (2010).
Blouin, M. S. DNA-based methods for pedigree reconstruction and kinship analysis in natural populations. Trends Ecol. Evol. 18, 503–511 (2003).
Goddard, M. E. & Hayes, B. J. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nature Rev. Genet. 10, 381–391 (2009).
Lacy, R. C. in Conservation Genetics in the Age of Genomics (eds Amato, G., Ryder, O., Rosenbaum, H. & DeSalle, R.) 58–81 (Columbia Univ. Press, New York, 2009).
Araki, H., Cooper, B. & Blouin, M. S. Genetic effects of captive breeding cause a rapid, cumulative fitness decline in the wild. Science 318, 100–103 (2007). One of the first papers to demonstrate a reduction in fitness in wild populations caused by gene flow from captive populations.
Frankham, R. Genetic adaptation to captivity in species conservation programs. Mol. Ecol. 17, 325–333 (2008).
Schwartz, M. K., Luikart, G. & Waples, R. S. Genetic monitoring as a promising tool for conservation and management. Trends Ecol. Evol. 22, 25–33 (2007). A foundation paper that defined and organized the emerging field of genetic monitoring.
Tallmon, D. A., Luikart, G. & Waples, R. S. The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol. 19, 489–496 (2004).
Piertney, S. B. & Oliver, M. K. The evolutionary ecology of the major histocompatibility complex. Heredity 96, 7–21 (2006).
Holmes, G. D., James, E. A. & Hoffmann, A. A. Limitations to reproductive output and genetic rescue in populations of the rare shrub Grevillea repens (Proteaceae). Ann. Bot. 102, 1031–1041 (2008).
Pogson, G. H. & Fevolden, S. E. Natural selection and the genetic differentiation of coastal and Arctic populations of the Atlantic cod in northern Norway: a test involving nucleotide sequence variation at the pantophysin (PanI) locus. Mol. Ecol. 12, 63–74 (2003).
Wheat, C. Phosphoglucose isomerase (Pgi) performance and fitness effects among Arthropods and its potential role as an adaptive marker in conservation genetics. Conserv. Genet. 11, 387–397 (2010).
Barbour, R. C., Forster, L. G., Baker, S. C., Steane, D. A. & Potts, B. M. Biodiversity consequences of genetic variation in bark characteristics within a foundation tree species. Conserv. Biol. 23, 1146–1155 (2009).
Whitham, T. G. et al. Extending genomics to natural communities and ecosystems. Science 320, 492–495 (2008).
Crutsinger, G. M. et al. Plant genotypic diversity predicts community structure and governs an ecosystem process. Science 313, 966–968 (2006).
Hodges, E. et al. Genome-wide in situ exon capture for selective resequencing. Nature Genet. 39, 1522–1527 (2007).
Nowrousian, M. Next-generation sequencing techniques for eukaryotic microorganisms: sequencing-based solutions to biological problems. Eukaryot. Cell 2 Jul 2010 (doi:10.1128/EC.00123-10).
Li, R. et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 19, 1124–1132 (2009).
International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
Haussler, D. et al. Genome 10K: a proposal to obtain whole-genome sequence for 10000 vertebrate species. J. Hered. 100, 659–674 (2009).
Bossdorf, O., Richards, C. L. & Pigliucci, M. Epigenetics for ecologists. Ecol. Lett. 11, 106–115 (2008). A valuable consideration of the future application of epigenetics to understanding the ecology of natural populations.
Richards, C. L., Bossdorf, O. & Pigliucci, M. What role does heritable epigenetic variation play in phenotypic evolution? BioScience 60, 232–237 (2010).
Salmon, A., Ainouche, M. L. & Wendel, J. F. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol. Ecol. 14, 1163–1175 (2005).
Richards, C. L. et al. Plasticity in salt tolerance traits allows for invasion of novel habitat by Japanese knotweed s. l. (Fallopia japonica and F. bohemica, Polygonaceae). Am. J. Bot. 95, 931–942 (2008).
Allendorf, F. W. & Lundquist, L. L. Introduction: population biology, evolution, and control of invasive species. Conserv. Biol. 17, 24–30 (2003).
Coulson, T. et al. Estimating individual contributions to population growth: evolutionary fitness in ecological time. Proc. Biol. Sci. 273, 547–555 (2006).
Palsbøll, P. J., Berube, M. & Allendorf, F. W. Identification of management units using population genetic data. Trends Ecol. Evol. 22, 11–16 (2007).
Waples, R. S. Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. J. Hered. 89, 438–450 (1998). An important paper that considers how to interpret the low genetic differentiation observed between marine populations that are apparently demographically isolated.
Pampoulie, C. et al. The genetic structure of Atlantic cod (Gadus morhua) around Iceland: insight from microsatellites, the PanI locus, and tagging experiments. Can. J. Fish. Aquat. Sci. 63, 2660–2674 (2006).
Hemmer-Hansen, J., Nielsen, E. E., Frydenberg, J. & Loeschcke, V. Adaptive divergence in a high gene flow environment: Hsc70 variation in the European flounder (Platichthys flesus L.). Heredity 99, 592–600 (2007).
Lowe, W. H. & Allendorf, F. W. What can genetics tell us about population connectivity? Mol. Ecol. 19, 3038–3051 (2010).
Waples, R. S. & Gaggiotti, O. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol. Ecol. 15, 1419–1439 (2006). An extremely valuable paper that considers the fundamental problem of defining 'population' in population genetics.
Baird, N. A. et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3, e3376 (2008).
Perkel, J. SNP genotyping: six technologies that keyed a revolution. Nature Methods 5, 447–453 (2008).
Decker, J. E. et al. Resolving the evolution of extant and extinct ruminants with high-throughput phylogenomics. Proc. Natl Acad. Sci. USA 106, 18644–18649 (2009).
Acknowledgements
This article is based partially on work supported by the US National Science Foundation grants DEB 074218 to F.W.A. and G.L., and IOS 0843392 to P.A.H. G.L. also received support from the Walton Family Foundation and research grants PTDC/BIA-BDE/65625/2006 and PTDC/CVT/69438/2006 from the Portuguese Science Foundation. We thank D. E. Campton, R. Frankham, O. Gaggiotti, P. Hedrick, L. S. Mills, B. A. Payseur, K. M. Ramstad, M. K. Schwartz, P. Sunnucks and D. A. Tallmon for useful comments, and W. H. Lowe for endless EndNote tutoring to F.W.A.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figure S1
The ability to detect local adaptation depends on gametic disequilibrium between the genotyped markers and loci under selection. (PDF 295 kb)
Related links
Glossary
- Neutral locus
-
A locus that has no effect on adaptation because all genotypes have the same fitness.
- Inbreeding coefficient
-
The probability that two alleles in an individual are both descended from a single allele in an ancestor (that is, that they are 'identical-by-descent').
- Contig
-
An abbreviation for contiguous sequence; used to indicate a contiguous piece of DNA that is assembled from shorter overlapping sequence reads.
- Metagenomics
-
The study of the collective genomic material contained in an environmental sample of microorganisms, facilitated by high-throughput sequencing technology that allows the direct sequencing of heterogeneous samples.
- Endosymbiont
-
An organism that lives within the cells of a host organism.
- Inbreeding depression
-
The loss of vigour and fitness that is observed when genome-wide homozygosity is increased by inbreeding.
- Adaptation
-
Heritable changes in genotype or phenotype that result in increased fitness.
- Hybridization
-
Interbreeding of individuals from genetically distinct populations, regardless of the taxonomic status of the populations.
- Outbreeding depression
-
Reduced fitness of F1 or F2 individuals after a cross between two species or populations. It can result from genetic incompatibility or reduced adaptation to local environmental conditions.
- Effective population size
-
The size of the ideal population that would experience the same amount of genetic drift as the observed population.
- Outlier locus
-
A genome location (or marker or base pair) that shows behaviour or a pattern of variation that is extremely divergent from the rest of the genome (locus-specific effects), as revealed by simulations or statistical tests.
- F ST
-
A measure of population subdivision that indicates the proportion of genetic diversity found between populations relative to the amount within populations.
- Population bottleneck
-
A marked reduction in population size followed by the survival and expansion of a small random sample of the original population. It often results in the loss of genetic variation and more frequent matings among closely related individuals.
- Hierarchical Bayesian model
-
A Bayesian model in which the prior depends on another parameter that is not in the likelihood function and that can vary and have another prior.
- Haplotype
-
A set of genetic markers that are present on a single chromosome and that show complete or nearly complete gametic disequilibrium. They are inherited through generations without being changed by crossing-over or other recombination mechanisms.
- Admixture
-
The production of new genetic combinations in hybrid populations through recombination.
- Coalescent approach
-
A means of investigating the shared genealogical history of genes. A genealogy is constructed backwards in time starting with the present-day sample. Lineages coalesce when they have a common ancestor.
- Selection coefficient
-
A term that describes the difference in average fitness between genotypes when fitness is measured relative to the average fitness of one of the genotypes (known as the reference genotype).
- Metapopulation
-
A collection of populations of a species found in differing geographic locations and with restricted gene flow (exchange of genes) between the populations.
- Proportion of admixture
-
The proportion of alleles in a hybrid swarm or individual that comes from each of the hybridizing taxa.
- Epistasis
-
The dependency of the effects of alleles at one locus on the genotypes at other loci in the genome.
- Purging
-
The selective reduction in frequency of deleterious recessive alleles in small populations because the increase in homozygosity increases the ability of selection to act on recessive alleles.
- Identical-by-descent
-
An allele shared by two related individuals is said to be identical-by-descent if the allele is inherited from the same common ancestor.
- Gametic disequilibrium
-
A measure of whether alleles at two loci in a population occur in a non-random fashion.
- Type I and type II errors
-
Statistical errors in which a true null hypothesis is rejected (type I) or a false null hypothesis is not rejected (type II).
- Expressed sequence tag
-
A short DNA fragment (several hundred base pairs) produced by reverse transcription of mRNA into DNA.
- Phenology
-
The timing of periodic biological phenomena that are usually correlated with climatic conditions.
- Landscape genomics
-
The study of many markers, including markers in genes under selection, in spatially referenced samples collected across a landscape and often across selection gradients. It uses comparisons of adaptive and neutral variation to quantify the effects of landscape features and environmental variables on gene flow and spatial genetic variation.
- Evolutionarily significant unit
-
A classification of populations that have substantial reproductive isolation which has led to adaptive differences so that the population represents a significant evolutionary component of the species.
- Distinct population segment
-
A classification under the Endangered Species Act of the United States that allows for legal protection of populations that are distinct, relatively reproductively isolated and represent a significant evolutionary lineage to the species.
- Management unit
-
A local population that is managed as a unit owing to its demographic independence.
- Introgression
-
Gene flow between populations or species whose individuals hybridize.
- Heterosis
-
When hybrid individuals have greater fitness than either of the parental types.
- Marker-assisted selection
-
The use of molecular genetic markers to increase the response to selection in a population by the favouring of reproduction by individuals with a certain allele or genotype. The marker is closely linked to a quantitative trait locus.
- Genetic rescue
-
The recovery in the average fitness of individuals through increased gene flow into small populations, typically following a fitness reduction due to inbreeding depression.
- Chondrodystrophy
-
A genetically based skeletal disorder that affects the development of cartilage.
- Community genomics
-
The study of the effect of individual alleles or genotypes on the species composition, diversity or functioning of a community or ecosystem.
- Epigenetics
-
Changes in or gene expression caused by mechanisms other than changes in the underlying DNA sequence, such as DNA methylation and histone modifications.
- Vital rates
-
Demographic values that affect population growth (for example, age-specific survival, fecundity and age at first reproduction).
Rights and permissions
About this article
Cite this article
Allendorf, F., Hohenlohe, P. & Luikart, G. Genomics and the future of conservation genetics. Nat Rev Genet 11, 697–709 (2010). https://doi.org/10.1038/nrg2844
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg2844
This article is cited by
-
Fasta2Structure: a user-friendly tool for converting multiple aligned FASTA files to STRUCTURE format
BMC Bioinformatics (2024)
-
High-quality faba bean reference transcripts generated using PacBio and Illumina RNA-seq data
Scientific Data (2024)
-
Microsatellite and mtDNA-based exploration of inter-generic hybridization and patterns of genetic diversity in major carps of Punjab, Pakistan
Aquaculture International (2024)
-
Sympatric species of coral trout (Plectropomus) show contrasting patterns of genomic structure across isolated atoll reefs
Reviews in Fish Biology and Fisheries (2024)
-
Mitochondrial DNA and local ecological knowledge reveal two lineages of leatherback turtle on the beaches of Oaxaca, Mexico
Scientific Reports (2023)